1YDK
 
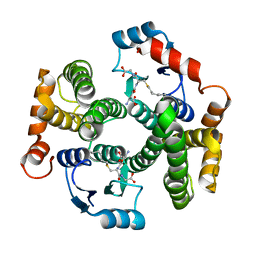 | | Crystal structure of the I219A mutant of human glutathione transferase A1-1 with S-hexylglutathione | | Descriptor: | Glutathione S-transferase A1, S-HEXYLGLUTATHIONE | | Authors: | Kuhnert, D.C, Sayed, Y, Mosebi, S, Sayed, M, Sewell, T, Dirr, H.W. | | Deposit date: | 2004-12-24 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Tertiary Interactions Stabilise the C-terminal Region of Human Glutathione Transferase A1-1: a Crystallographic and Calorimetric Study.
J.Mol.Biol., 349, 2005
|
|
4ALV
 
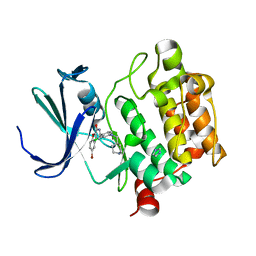 | | Benzofuropyrimidinone Inhibitors of Pim-1 | | Descriptor: | 8-bromo-2-{2-chloro-4-[(piperidin-4-ylmethyl)amino]phenyl}[1]benzofuro[3,2-d]pyrimidin-4(3H)-one, IMIDAZOLE, SERINE/THREONINE-PROTEIN KINASE PIM-1 | | Authors: | Stout, T.J, Adams, L. | | Deposit date: | 2012-03-05 | | Release date: | 2013-01-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | The Design, Synthesis, and Biological Evaluation of Pim Kinase Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2FF0
 
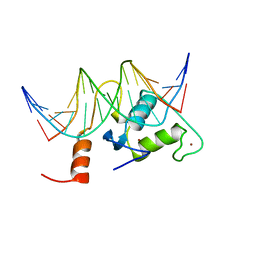 | | Solution Structure of Steroidogenic Factor 1 DNA Binding Domain Bound to its Target Sequence in the Inhibin alpha-subunit Promoter | | Descriptor: | CTGTGGCCCTGAGCC, GGCTCAGGGCCACAG, Steroidogenic factor 1, ... | | Authors: | Little, T.H, Zhang, Y, Matulis, C.K, Weck, J, Zhang, Z, Ramachandran, A, Mayo, K.E, Radhakrishnan, I. | | Deposit date: | 2005-12-17 | | Release date: | 2006-04-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific deoxyribonucleic Acid (DNA) recognition by steroidogenic factor 1: a helix at the carboxy terminus of the DNA binding domain is necessary for complex stability.
Mol.Endocrinol., 20, 2006
|
|
1Q94
 
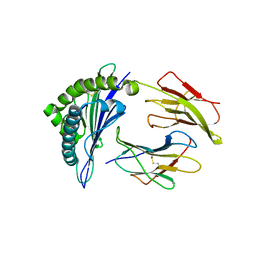 | | Structures of HLA-A*1101 in complex with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle anchor residue | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-11 alpha chain, ... | | Authors: | Li, L, McNicholl, J.M, Bouvier, M. | | Deposit date: | 2003-08-22 | | Release date: | 2004-06-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of HLA-A*1101 complexed with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle, secondary anchor residue.
J.Immunol., 172, 2004
|
|
2FF1
 
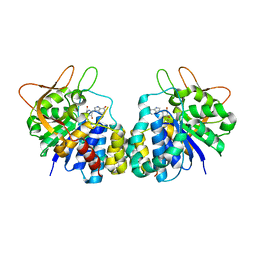 | | Crystal structure of Trypanosoma vivax nucleoside hydrolase soaked with ImmucillinH | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, IAG-nucleoside hydrolase | | Authors: | Versees, W, Barlow, J, Steyaert, J. | | Deposit date: | 2005-12-18 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Transition-state Complex of the Purine-specific Nucleoside Hydrolase of T.vivax: Enzyme Conformational Changes and Implications for Catalysis.
J.Mol.Biol., 359, 2006
|
|
2PF2
 
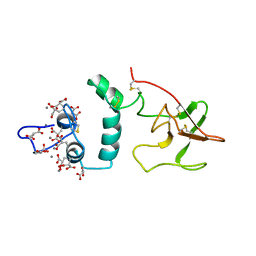 | | THE CA+2 ION AND MEMBRANE BINDING STRUCTURE OF THE GLA DOMAIN OF CA-PROTHROMBIN FRAGMENT 1 | | Descriptor: | CALCIUM ION, PROTHROMBIN FRAGMENT 1 | | Authors: | Soriano-Garcia, M, Padmanabhan, K, De vos, A.M, Tulinsky, A. | | Deposit date: | 1991-12-08 | | Release date: | 1994-01-31 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Ca2+ ion and membrane binding structure of the Gla domain of Ca-prothrombin fragment 1.
Biochemistry, 31, 1992
|
|
3KT2
 
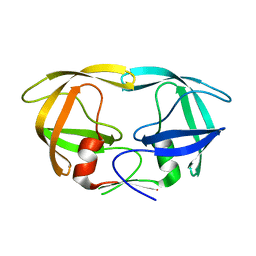 | | Crystal Structure of N88D mutant HIV-1 Protease | | Descriptor: | Protease | | Authors: | Bihani, S.C, Das, A, Prashar, V, Ferrer, J.L, Hosur, M.V. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Resistance mechanism revealed by crystal structures of unliganded nelfinavir-resistant HIV-1 protease non-active site mutants N88D and N88S.
Biochem.Biophys.Res.Commun., 389, 2009
|
|
4H6J
 
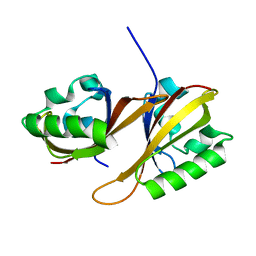 | | Identification of Cys 255 in HIF-1 as a novel site for development of covalent inhibitors of HIF-1 /ARNT PasB domain protein-protein interaction. | | Descriptor: | ARYL HYDROCARBON NUCLEAR TRANSLOCATOR, HYPOXIA INDUCIBLE FACTOR 1-ALPHA | | Authors: | Cardoso, R, Love, R.A, Nilsson, C, Bergqvist, S, Nowlin, D, Yan, J, Liu, K, Zhu, J, Chen, P, Deng, Y.-L, Dyson, H.J, Greig, M.J, Brooun, A. | | Deposit date: | 2012-09-19 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Identification of Cys255 in HIF-1 alpha as a novel site for development of covalent inhibitors of HIF-1 alpha /ARNT PasB domain protein-protein interaction.
Protein Sci., 21, 2012
|
|
1V3L
 
 | | Crystal structure of F283L mutant cyclodextrin glycosyltransferase complexed with a pseudo-tetraose derived from acarbose | | Descriptor: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-beta-D-galactopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | Authors: | Kanai, R, Haga, K, Akiba, T, Yamane, K, Harata, K. | | Deposit date: | 2003-11-03 | | Release date: | 2004-08-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of Phe283 in enzymatic reaction of cyclodextrin glycosyltransferase from alkalophilic Bacillus sp.1011: Substrate binding and arrangement of the catalytic site
PROTEIN SCI., 13, 2004
|
|
5OA2
 
 | | Crystal structure of ScGas2 in complex with compound 8 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-beta-glucanosyltransferase GAS2, SULFATE ION, ... | | Authors: | Delso, I, Valero-Gonzalez, J, Gomollon-Bel, F, Castro-Lopez, J, Fang, W, Navratilova, I, Van Aalten, D, Tejero, T, Merino, P, Hurtado-Guerrero, R. | | Deposit date: | 2017-06-20 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Inhibitors against Fungal Cell Wall Remodeling Enzymes.
ChemMedChem, 13, 2018
|
|
1QGI
 
 | | CHITOSANASE FROM BACILLUS CIRCULANS | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (CHITOSANASE), SULFATE ION | | Authors: | Saito, J, Kita, A, Higuchi, Y, Nagata, Y, Ando, A, Miki, K. | | Deposit date: | 1999-04-28 | | Release date: | 1999-10-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of chitosanase from Bacillus circulans MH-K1 at 1.6-A resolution and its substrate recognition mechanism.
J.Biol.Chem., 274, 1999
|
|
2F3K
 
 | | Substrate envelope and drug resistance: crystal structure of r01 in complex with wild-type hiv-1 protease | | Descriptor: | (3S,4AS,8AS)-N-(TERT-BUTYL)-2-[(3S)-3-({3-(METHYLSULFONYL)-N-[(PYRIDIN-3-YLOXY)ACETYL]-L-VALYL}AMINO)-2-OXO-4-PHENYLBUTYL]DECAHYDROISOQUINOLINE-3-CARBOXAMIDE, PHOSPHATE ION, Protease | | Authors: | Prabu-Jeyabalan, M, King, N.M, Nalivaika, E.A, Heilek-Snyder, G, Cammack, N, Schiffer, C.A. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Substrate envelope and drug resistance: crystal structure of RO1 in complex with wild-type human immunodeficiency virus type 1 protease.
Antimicrob.Agents Chemother., 50, 2006
|
|
3WRY
 
 | | Crystal structure of helicase complex 2 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Matsumura, H, Katoh, E. | | Deposit date: | 2014-02-27 | | Release date: | 2014-08-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the recognition-evasion arms race between Tomato mosaic virus and the resistance gene Tm-1
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5XC2
 
 | | Crystal structure of GH family 81 beta-1,3-glucanase from Rhizomucr miehei complexed with laminarihexaose | | Descriptor: | Endo-beta-1,3-glucanase, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose, ... | | Authors: | Qin, Z, Yang, S, Peng, Z, Yan, Q, Jiang, Z. | | Deposit date: | 2017-03-22 | | Release date: | 2018-03-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Catalytic mechanism of glycoside hydrolase family 81 beta-1,3-glucanase
To Be Published
|
|
3KT5
 
 | | Crystal Structure of N88S mutant HIV-1 Protease | | Descriptor: | Protease | | Authors: | Bihani, S.C, Das, A, Prashar, V, Ferrer, J.L, Hosur, M.V. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Resistance mechanism revealed by crystal structures of unliganded nelfinavir-resistant HIV-1 protease non-active site mutants N88D and N88S.
Biochem.Biophys.Res.Commun., 389, 2009
|
|
1E1Y
 
 | | Flavopiridol inhibits glycogen phosphorylase by binding at the inhibitor site | | Descriptor: | 2-(2-CHLORO-PHENYL)-5,7-DIHYDROXY-8-(3-HYDROXY-1-METHYL-PIPERIDIN-4-YL)-4H-BENZOPYRAN-4-ONE, GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, ... | | Authors: | Oikonomakos, N.G, Zographos, S.E, Skamnaki, V.T, Tsitsanou, K.E, Johnson, L.N. | | Deposit date: | 2000-05-11 | | Release date: | 2000-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Flavopiridol Inhibits Glycogen Phosphorylase by Binding at the Inhibitor Site
J.Biol.Chem., 275, 2000
|
|
1E3O
 
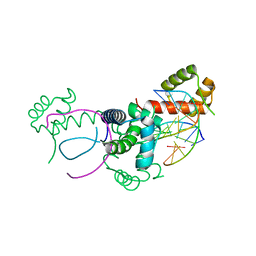 | | Crystal structure of Oct-1 POU dimer bound to MORE | | Descriptor: | 5'-D(*AP*TP*GP*CP*AP*TP*GP*AP*GP*GP*A)-3', 5'-D(*TP*CP*CP*TP*CP*AP*TP*GP*CP*AP*T)-3', OCTAMER-BINDING TRANSCRIPTION FACTOR 1 | | Authors: | Remenyi, A, Tomilin, A, Pohl, E, Schoeler, H, Wilmanns, M. | | Deposit date: | 2000-06-20 | | Release date: | 2001-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Differential Dimer Activities of the Transcription Factor Oct-1 by DNA-Induced Interface Swapping
Mol.Cell, 8, 2001
|
|
2YCW
 
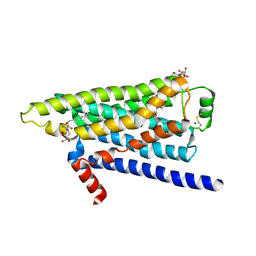 | | TURKEY BETA1 ADRENERGIC RECEPTOR WITH STABILISING MUTATIONS AND BOUND ANTAGONIST CARAZOLOL | | Descriptor: | (2S)-1-(9H-Carbazol-4-yloxy)-3-(isopropylamino)propan-2-ol, BETA-1 ADRENERGIC RECEPTOR, HEGA-10, ... | | Authors: | Moukhametzianov, R, Warne, T, Edwards, P.C, Serrano-Vega, M.J, Leslie, A.G.W, Tate, C.G, Schertler, G.F.X. | | Deposit date: | 2011-03-17 | | Release date: | 2011-06-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two Distinct Conformations of Helix 6 Observed in Antagonist-Bound Structures of a Beta-1- Adrenergic Receptor.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2FF2
 
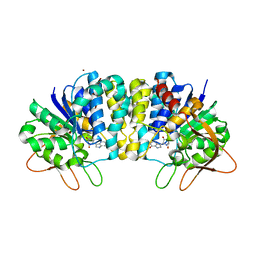 | | Crystal structure of Trypanosoma vivax nucleoside hydrolase co-crystallized with ImmucillinH | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, CALCIUM ION, IAG-nucleoside hydrolase, ... | | Authors: | Versees, W, Barlow, J, Steyaert, J. | | Deposit date: | 2005-12-18 | | Release date: | 2006-05-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Transition-state Complex of the Purine-specific Nucleoside Hydrolase of T.vivax: Enzyme Conformational Changes and Implications for Catalysis.
J.Mol.Biol., 359, 2006
|
|
2PGN
 
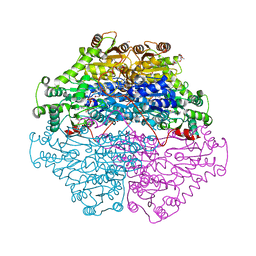 | | The crystal structure of FAD and ThDP-dependent Cyclohexane-1,2-dione Hydrolase in Complex with Cyclohexane-1,2-dione | | Descriptor: | CYCLOHEXANE-1,2-DIONE, Cyclohexane-1,2-dione Hydrolase (Cdh), FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fraas, S, Warkentin, E, Ermler, U. | | Deposit date: | 2007-04-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The crystal structure of FAD and ThDP-dependent Cyclohexane-1,2-dione Hydrolase in Complex with Cyclohexane-1,2-dione
To be Published
|
|
1J5O
 
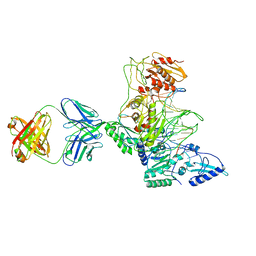 | | CRYSTAL STRUCTURE OF MET184ILE MUTANT OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH DOUBLE STRANDED DNA TEMPLATE-PRIMER | | Descriptor: | 5'-D(*AP*TP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*C)-3', 5'-D(*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3', ANTIBODY (HEAVY CHAIN), ... | | Authors: | Sarafianos, S.G, Das, K, Arnold, E. | | Deposit date: | 2002-05-24 | | Release date: | 2002-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1RS2
 
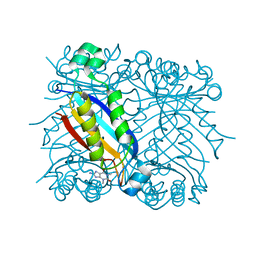 | | DHNA complex with 8-Amino-1,3-dimethyl-3,7-dihydropurine-2,6-dione | | Descriptor: | 8-AMINO-1,3-DIMETHYL-3,7-DIHYDROPURINE-2,6-DIONE, Dihydroneopterin aldolase | | Authors: | Sanders, W.J, Nienaber, V.L, Lerner, C.G, McCall, J.O, Merrick, S.M, Swanson, S.J, Harlan, J.E, Stoll, V.S, Stamper, G.F, Betz, S.F, Condroski, K.R, Meadows, R.P, Severin, J.M, Walter, K.A, Magdalinos, P, Jakob, C.G, Wagner, R, Beutel, B.A. | | Deposit date: | 2003-12-09 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Discovery of Potent Inhibitors of Dihydroneopterin Aldolase Using CrystaLEAD High-Throughput X-ray Crystallographic Screening and Structure-Directed Lead Optimization.
J.Med.Chem., 47, 2004
|
|
1RWM
 
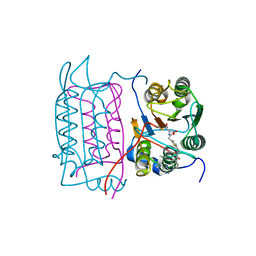 | | Crystal structure of human caspase-1 in complex with 4-oxo-3-[2-(5-{[4-(quinoxalin-2-ylamino)-benzoylamino]-methyl}-thiophen-2-yl)-acetylamino]-pentanoic acid | | Descriptor: | 4-OXO-3-[2-(5-{[4-(QUINOXALIN-2-YLAMINO)-BENZOYLAMINO]-METHYL}-THIOPHEN-2-YL)-ACETYLAMINO]-PENTANOIC ACID, Interleukin-1 beta convertase | | Authors: | Romanowski, M.J, Waal, N.D, Fahr, B.T, O'Brien, T. | | Deposit date: | 2003-12-16 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of caspase-1 inhibitors derived from Tethering.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
4GO6
 
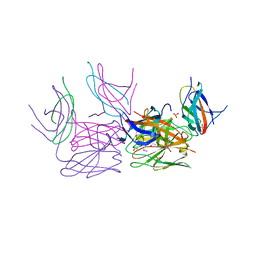 | | Crystal structure of HCF-1 self-association sequence 1 | | Descriptor: | HCF C-terminal chain 1, HCF N-terminal chain 1, SULFATE ION | | Authors: | Park, J, Lammers, F, Herr, W, Song, J. | | Deposit date: | 2012-08-18 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | HCF-1 self-association via an interdigitated Fn3 structure facilitates transcriptional regulatory complex formation
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3B7E
 
 | | Neuraminidase of A/Brevig Mission/1/1918 H1N1 strain in complex with zanamivir | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Xu, X, Zhu, X, Wilson, I.A. | | Deposit date: | 2007-10-30 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural characterization of the 1918 influenza virus H1N1 neuraminidase
J.Virol., 82, 2008
|
|
