8E92
 
 | | D-cycloserine and glutamate bound Human GluN1a-GluN2C NMDA receptor in intact conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E93
 
 | | D-cycloserine and glutamate bound Human GluN1a-GluN2C NMDA receptor in splayed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
8E98
 
 | | D-cycloserine and glutamate bound Human GluN1a-GluN2C NMDA receptor in nanodisc - intact conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Chou, T.-H, Furukawa, H. | | Deposit date: | 2022-08-26 | | Release date: | 2022-12-07 | | Last modified: | 2022-12-14 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural insights into assembly and function of GluN1-2C, GluN1-2A-2C, and GluN1-2D NMDARs.
Mol.Cell, 82, 2022
|
|
2JA1
 
 | | Thymidine kinase from B. cereus with TTP bound as phosphate donor. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, THYMIDINE KINASE, THYMIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosinska, U, Carnrot, C, Sandrini, M.P.B, Clausen, A.R, Wang, L, Piskur, J, Eriksson, S, Eklund, H. | | Deposit date: | 2006-11-17 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Studies of Thymidine Kinases from Bacillus Anthracis and Bacillus Cereus Provide Insights Into Quaternary Structure and Conformational Changes Upon Substrate Binding
FEBS J., 274, 2007
|
|
2OX3
 
 | | R-state, PEP and Fru-6-P-bound, Escherichia coli fructose-1,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase, PHOSPHOENOLPYRUVATE | | Authors: | Hines, J.K, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-02-19 | | Release date: | 2007-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of activated fructose-1,6-bisphosphatase from Escherichia coli. Coordinate regulation of bacterial metabolism and the conservation of the R-state.
J.Biol.Chem., 282, 2007
|
|
2HBS
 
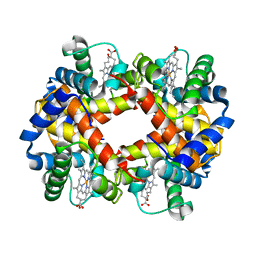 | | THE HIGH RESOLUTION CRYSTAL STRUCTURE OF DEOXYHEMOGLOBIN S | | Descriptor: | HEMOGLOBIN S (DEOXY), ALPHA CHAIN, BETA CHAIN, ... | | Authors: | Harrington, D.J, Adachi, K, Royer Junior, W.E. | | Deposit date: | 1997-05-06 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The high resolution crystal structure of deoxyhemoglobin S.
J.Mol.Biol., 272, 1997
|
|
1D8W
 
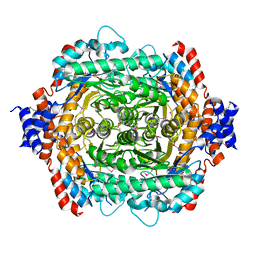 | | L-RHAMNOSE ISOMERASE | | Descriptor: | L-RHAMNOSE ISOMERASE, ZINC ION | | Authors: | Korndorfer, I.P, Matthews, B.W. | | Deposit date: | 1999-10-26 | | Release date: | 2000-09-27 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of rhamnose isomerase from Escherichia coli and its relation with xylose isomerase illustrates a change between inter and intra-subunit complementation during evolution.
J.Mol.Biol., 300, 2000
|
|
1A16
 
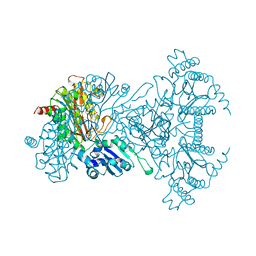 | | AMINOPEPTIDASE P FROM E. COLI WITH THE INHIBITOR PRO-LEU | | Descriptor: | AMINOPEPTIDASE P, LEUCINE, MANGANESE (II) ION, ... | | Authors: | Wilce, M.C, Bond, C.S, Lilley, P.E, Dixon, N.E, Freeman, H.C, Guss, J.M. | | Deposit date: | 1997-12-22 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4Y3O
 
 | | Crystal structure of Ribosomal oxygenase NO66 in complex with substrate Rpl8 peptide and Ni(II) and cofactor N-oxalyglycine | | Descriptor: | ACETATE ION, Bifunctional lysine-specific demethylase and histidyl-hydroxylase NO66, GLYCEROL, ... | | Authors: | Wang, C, Zhang, Q, Zang, J. | | Deposit date: | 2015-02-10 | | Release date: | 2015-10-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the JmjC domain-containing protein NO66 complexed with ribosomal protein Rpl8.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WK6
 
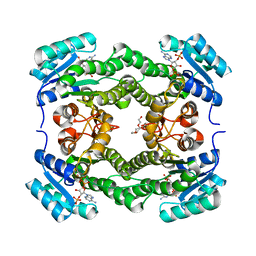 | | Crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase (FabG) (G141A) from Vibrio cholerae in complex with NADPH | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PENTAETHYLENE GLYCOL | | Authors: | Hou, J, Zheng, H, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-10-01 | | Release date: | 2014-11-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
8OJJ
 
 | | Cryo-EM structure of the DnaD-NTD tetramer | | Descriptor: | DNA replication protein DnaD | | Authors: | Winterhalter, C, Pelliciari, S, Cronin, N, Costa, T.R.D, Murray, H, Ilangovan, A. | | Deposit date: | 2023-03-24 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (5.47 Å) | | Cite: | The DNA replication initiation protein DnaD recognises a specific strand of the Bacillus subtilis chromosome origin.
Nucleic Acids Res., 51, 2023
|
|
6GNM
 
 | | Crystal Structure Of Sea Bream Transthyretin in complex with 2,2',4,4'-tetrahydroxybenzophenone (BP2) | | Descriptor: | Transthyretin, bis(2,4-dihydroxyphenyl)methanone | | Authors: | Grundstrom, C, Zhang, J, Olofsson, A, Andersson, P.L, Sauer-Eriksson, A.E. | | Deposit date: | 2018-05-31 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Interspecies Variation between Fish and Human Transthyretins in Their Binding of Thyroid-Disrupting Chemicals.
Environ. Sci. Technol., 52, 2018
|
|
1H2B
 
 | | Crystal Structure of the Alcohol Dehydrogenase from the Hyperthermophilic Archaeon Aeropyrum pernix at 1.65A Resolution | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE (ACIDIC FORM), OCTANOIC ACID (CAPRYLIC ACID), ... | | Authors: | Guy, J.E, Isupov, M.N, Littlechild, J.A. | | Deposit date: | 2002-08-02 | | Release date: | 2003-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structure of an alcohol dehydrogenase from the hyperthermophilic archaeon Aeropyrum pernix.
J.Mol.Biol., 331, 2003
|
|
4ZHE
 
 | | Crystal structure of the SeMet substituted Topless related protein 2 (TPR2) N-terminal domain (1-209) from rice | | Descriptor: | ASPR2 protein | | Authors: | Ke, J, Ma, H, Gu, X, Brunzelle, J.S, Xu, H.E, Melcher, K. | | Deposit date: | 2015-04-24 | | Release date: | 2015-08-05 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for recognition of diverse transcriptional repressors by the TOPLESS family of corepressors.
Sci Adv, 1, 2015
|
|
1AYY
 
 | | GLYCOSYLASPARAGINASE | | Descriptor: | GLYCOSYLASPARAGINASE | | Authors: | Van Roey, P, Xuan, J. | | Deposit date: | 1997-11-12 | | Release date: | 1998-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Crystal structure of glycosylasparaginase from Flavobacterium meningosepticum.
Protein Sci., 7, 1998
|
|
2L0J
 
 | | Solid State NMR structure of the M2 proton channel from Influenza A Virus in hydrated lipid bilayer | | Descriptor: | Matrix protein 2 | | Authors: | Sharma, M, Yi, M, Dong, H, Qin, H, Peterson, E, Busath, D.D, Zhou, H.X, Cross, T.A. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Insight into the mechanism of the influenza a proton channel from a structure in a lipid bilayer.
Science, 330, 2010
|
|
7MQL
 
 | | AAC(3)-IIIa in complex with CoA and neomycin | | Descriptor: | Aminoglycoside N(3)-acetyltransferase III, COENZYME A, FORMIC ACID, ... | | Authors: | Zielinski, M, Berghuis, A.M. | | Deposit date: | 2021-05-05 | | Release date: | 2022-07-06 | | Last modified: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural elucidation of substrate-bound aminoglycoside acetyltransferase (3)-IIIa.
Plos One, 17, 2022
|
|
1DE5
 
 | | L-RHAMNOSE ISOMERASE | | Descriptor: | L-RHAMNITOL, L-RHAMNOSE ISOMERASE, ZINC ION | | Authors: | Korndorfer, I.P, Fessner, W.D, Matthews, B.W. | | Deposit date: | 1999-11-12 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of rhamnose isomerase from Escherichia coli and its relation with xylose isomerase illustrates a change between inter and intra-subunit complementation during evolution.
J.Mol.Biol., 300, 2000
|
|
1DE6
 
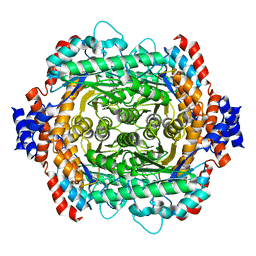 | | L-RHAMNOSE ISOMERASE | | Descriptor: | L-RHAMNOSE, L-RHAMNOSE ISOMERASE, MANGANESE (II) ION, ... | | Authors: | Korndorfer, I.P, Fessner, W.D, Matthews, B.W. | | Deposit date: | 1999-11-13 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of rhamnose isomerase from Escherichia coli and its relation with xylose isomerase illustrates a change between inter and intra-subunit complementation during evolution.
J.Mol.Biol., 300, 2000
|
|
1IOF
 
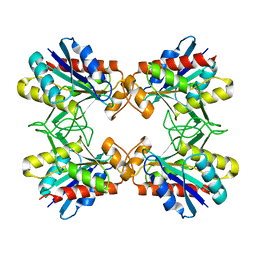 | | X-RAY CRYSTALLINE STRUCTURES OF PYRROLIDONE CARBOXYL PEPTIDASE FROM A HYPERTHERMOPHILE, PYROCOCCUS FURIOSUS, AND ITS CYS-FREE MUTANT | | Descriptor: | PYRROLIDONE CARBOXYL PEPTIDASE | | Authors: | Tanaka, H, Chinami, M, Ota, M, Tsukihara, T, Yutani, K. | | Deposit date: | 2001-03-09 | | Release date: | 2001-03-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystalline structures of pyrrolidone carboxyl peptidase from a hyperthermophile, Pyrococcus furiosus, and its cys-free mutant.
J.Biochem., 130, 2001
|
|
1IOI
 
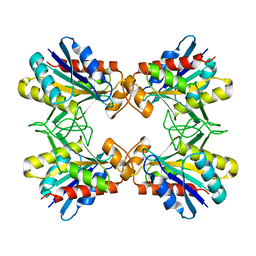 | | x-ray crystalline structures of pyrrolidone carboxyl peptidase from a hyperthermophile, pyrococcus furiosus, and its cys-free mutant | | Descriptor: | PYRROLIDONE CARBOXYL PEPTIDASE | | Authors: | Tanaka, H, Chinami, M, Ota, M, Tsukihara, T, Yutani, K. | | Deposit date: | 2001-03-09 | | Release date: | 2001-03-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray crystalline structures of pyrrolidone carboxyl peptidase from a hyperthermophile, Pyrococcus furiosus, and its cys-free mutant.
J.Biochem., 130, 2001
|
|
2OWZ
 
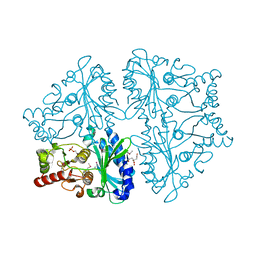 | | R-state, citrate and Fru-6-P-bound Escherichia coli fructose-1,6-bisphosphatase | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, CITRIC ACID, Fructose-1,6-bisphosphatase | | Authors: | Hines, J.K, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-02-17 | | Release date: | 2007-03-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structures of activated fructose-1,6-bisphosphatase from Escherichia coli. Coordinate regulation of bacterial metabolism and the conservation of the R-state.
J.Biol.Chem., 282, 2007
|
|
1J7S
 
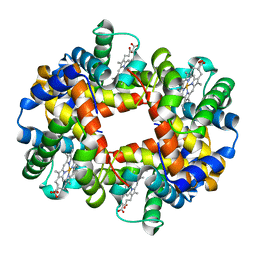 | | Crystal Structure of deoxy HbalphaYQ, a mutant of HbA | | Descriptor: | Hemoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Miele, A.E, Draghi, F, Arcovito, A, Bellelli, A, Brunori, M, Travaglini-Allocatelli, C, Vallone, B. | | Deposit date: | 2001-05-18 | | Release date: | 2002-02-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Control of heme reactivity by diffusion: structural basis and functional characterization in hemoglobin mutants.
Biochemistry, 40, 2001
|
|
2QRV
 
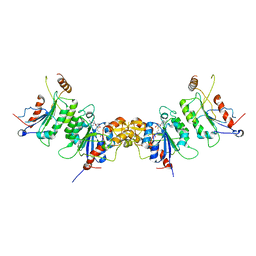 | | Structure of Dnmt3a-Dnmt3L C-terminal domain complex | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Jia, D, Cheng, X. | | Deposit date: | 2007-07-29 | | Release date: | 2007-12-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation.
Nature, 449, 2007
|
|
7PZL
 
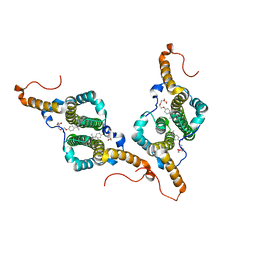 | | HBc-F97L premature secretion phenotype | | Descriptor: | Capsid protein, FRAGMENT OF TRITON X-100 | | Authors: | Makbul, C, Boettcher, B. | | Deposit date: | 2021-10-12 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Binding of a Pocket Factor to Hepatitis B Virus Capsids Changes the Rotamer Conformation of Phenylalanine 97.
Viruses, 13, 2021
|
|
