4GL5
 
 | |
1TAQ
 
 | | STRUCTURE OF TAQ DNA POLYMERASE | | Descriptor: | 2-O-octyl-beta-D-glucopyranose, TAQ DNA POLYMERASE, ZINC ION | | Authors: | Kim, Y, Eom, S.H, Wang, J, Lee, D.-S, Suh, S.W, Steitz, T.A. | | Deposit date: | 1996-06-04 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Thermus aquaticus DNA polymerase.
Nature, 376, 1995
|
|
3QU8
 
 | | Crystal structure of a human cytochrome P450 2B6 (Y226H/K262R) in complex with the inhibitor 4-(4-Nitrobenzyl)pyridine. | | Descriptor: | 4-(4-nitrobenzyl)pyridine, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Shah, M.B, Pascual, J, Stout, C.D, Halpert, J.R. | | Deposit date: | 2011-02-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Cytochrome P450 2B6 Bound to 4-Benzylpyridine and 4-(4-Nitrobenzyl)pyridine: Insight into Inhibitor Binding and Rearrangement of Active Site Side Chains.
Mol.Pharmacol., 80, 2011
|
|
1TIK
 
 | | CRYSTAL STRUCTURE OF ACYL CARRIER PROTEIN PHOSPHODIESTERASE | | Descriptor: | Acyl carrier protein phosphodiesterase, SULFATE ION | | Authors: | Rajashankar, K.R, Kniewel, R, Solorzano, V, Lima, C.D, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2004-06-02 | | Release date: | 2004-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRYSTAL STRUCTURE OF ACYL CARRIER PROTEIN PHOSPHODIESTERASE
To be Published
|
|
7M30
 
 | |
7M5U
 
 | | Crystal structure of human MPP8 chromodomain in complex with peptidomimetic ligand UNC5246 | | Descriptor: | M-phase phosphoprotein 8, UNC5246 | | Authors: | Budziszewski, G.R, McGinty, R.K, Waybright, J.M, Norris, J.L, James, L.I. | | Deposit date: | 2021-03-24 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A Peptidomimetic Ligand Targeting the Chromodomain of MPP8 Reveals HRP2's Association with the HUSH Complex.
Acs Chem.Biol., 16, 2021
|
|
4G2G
 
 | | Crystal structure of Mycobacterium tuberculosis CYP121 in complex with 4,4'-(1H-1,2,3-triazole-1,5-diyl)diphenol | | Descriptor: | 4,4'-(1H-1,2,3-triazole-1,5-diyl)diphenol, Cytochrome P450 121, DIMETHYL SULFOXIDE, ... | | Authors: | Hudson, S.A. | | Deposit date: | 2012-07-12 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
7MSJ
 
 | | The crystal structure of mouse HVEM | | Descriptor: | SULFATE ION, Tumor necrosis factor receptor superfamily member 14 | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
7MSG
 
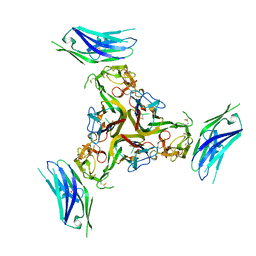 | | The crystal structure of LIGHT in complex with HVEM and CD160 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CD160 antigen, soluble form,Tumor necrosis factor receptor superfamily member 14, ... | | Authors: | Liu, W, Ramagopal, U, Garrett-Thompson, S.C, Fedorov, E, Bonanno, J.B, Almo, S.C. | | Deposit date: | 2021-05-11 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | HVEM structures and mutants reveal distinct functions of binding to LIGHT and BTLA/CD160.
J.Exp.Med., 218, 2021
|
|
7MK7
 
 | | Augmentor domain of augmentor-beta | | Descriptor: | ALK and LTK ligand 1,Maltodextrin-binding protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Krimmer, S.G, Reshetnyak, A.V, Puleo, D.E, Schlessinger, J. | | Deposit date: | 2021-04-21 | | Release date: | 2021-11-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42815185 Å) | | Cite: | Structural basis for ligand reception by anaplastic lymphoma kinase.
Nature, 600, 2021
|
|
1RK6
 
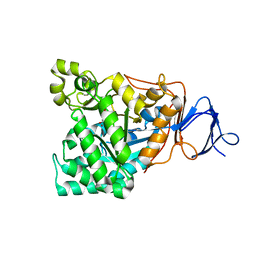 | | The enzyme in complex with 50mM CdCl2 | | Descriptor: | ACETATE ION, CADMIUM ION, D-aminoacylase, ... | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
1B74
 
 | | GLUTAMATE RACEMASE FROM AQUIFEX PYROPHILUS | | Descriptor: | D-GLUTAMINE, GLUTAMATE RACEMASE | | Authors: | Hwang, K.Y, Cho, C.S, Kim, S.S, Yu, Y.G, Cho, Y. | | Deposit date: | 1999-01-27 | | Release date: | 2000-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of glutamate racemase from Aquifex pyrophilus.
Nat.Struct.Biol., 6, 1999
|
|
1RJR
 
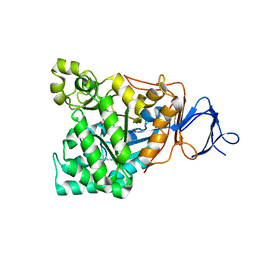 | | The crystal structure of the D-aminoacylase D366A mutant in complex with 100mM ZnCl2 | | Descriptor: | ACETATE ION, D-aminoacylase, ZINC ION | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
4G1X
 
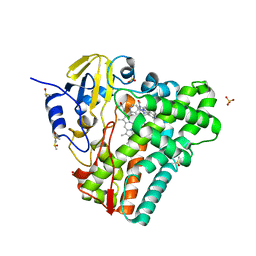 | | Crystal structure of Mycobacterium tuberculosis CYP121 in complex with 4-(1H-1,2,4-triazol-1-yl)quinolin-6-amine | | Descriptor: | 4-(1H-1,2,4-triazol-1-yl)quinolin-6-amine, Cytochrome P450 121, DIMETHYL SULFOXIDE, ... | | Authors: | Hudson, S.A. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
7MSL
 
 | |
4G48
 
 | | Structure of CYP121 in complex with 4-(4-phenoxy-1H-pyrazol-3-yl)benzene-1,3-diol | | Descriptor: | 4-(4-phenoxy-1H-pyrazol-3-yl)benzene-1,3-diol, Cytochrome P450 121, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hudson, S.A, McLean, K.J, Surade, S, Yang, Y.-Q, Leys, D, Ciulli, A, Munro, A.W, Abell, C. | | Deposit date: | 2012-07-16 | | Release date: | 2012-09-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Application of Fragment Screening and Merging to the Discovery of Inhibitors of the Mycobacterium tuberculosis Cytochrome P450 CYP121
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
6AB5
 
 | | Cryo-EM structure of T=1 Penaeus vannamei nodavirus | | Descriptor: | Capsid protein | | Authors: | Chen, N.C, Miyazaki, N, Yoshimura, M, Guan, H.H, Lin, C.C, Iwasaki, K, Chen, C.J. | | Deposit date: | 2018-07-20 | | Release date: | 2019-03-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The atomic structures of shrimp nodaviruses reveal new dimeric spike structures and particle polymorphism.
Commun Biol, 2, 2019
|
|
4G2N
 
 | | Crystal structure of putative D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding from Polaromonas sp. JS6 66 | | Descriptor: | CHLORIDE ION, D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding, ... | | Authors: | Malashkevich, V.N, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Zenchek, W, Love, J, Fiser, A, Khafizov, K, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-12 | | Release date: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative D-isomer specific 2-hydroxyacid dehydrogenase, NAD-binding from Polaromonas sp. JS6 66
To be Published
|
|
3QOA
 
 | | Crystal structure of a human cytochrome P450 2B6 (Y226H/K262R) in complex with the inhibitor 4-Benzylpyridine. | | Descriptor: | 4-benzylpyridine, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Shah, M.B, Pascual, J, Stout, C.D, Halpert, J.R. | | Deposit date: | 2011-02-09 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Cytochrome P450 2B6 Bound to 4-Benzylpyridine and 4-(4-Nitrobenzyl)pyridine: Insight into Inhibitor Binding and Rearrangement of Active Site Side Chains.
Mol.Pharmacol., 80, 2011
|
|
7MNJ
 
 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 145-673) | | Descriptor: | E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Patke, A, Dasso, M, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNK
 
 | | Crystal structure of the tetramerization element of NUP358/RanBP2 (residues 805-832) | | Descriptor: | 1,2-ETHANEDIOL, E3 SUMO-protein ligase RanBP2, SULFATE ION | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNO
 
 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 1-752) I656V mutant in complex with Fab fragment | | Descriptor: | Antibody Fab14 Heavy Chain, Antibody Fab14 Light Chain, E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (6.73 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNL
 
 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 1-752) in complex with Fab fragment | | Descriptor: | Antibody Fab14 Heavy Chain, Antibody Fab14 Light Chain, E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.95 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNN
 
 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 1-752) T653I mutant in complex with Fab fragment | | Descriptor: | Antibody Fab14 Heavy Chain, Antibody Fab14 Light Chain, E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (6.7 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNI
 
 | | Crystal structure of the N-terminal domain of NUP88 in complex with NUP98 C-terminal Autoproteolytic Domain | | Descriptor: | Nuclear pore complex protein Nup88, Nuclear pore complex protein Nup98 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
