6N8J
 
 | | Cryo-EM structure of late nuclear (LN) pre-60S ribosomal subunit | | Descriptor: | 5.8S rRNA, 5S rRNA, 60S ribosomal protein L11-A, ... | | Authors: | Zhou, Y, Musalgaonkar, S, Johnson, A.W, Taylor, D.W. | | Deposit date: | 2018-11-29 | | Release date: | 2019-03-13 | | Last modified: | 2020-01-08 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome.
Nat Commun, 10, 2019
|
|
5YWB
 
 | |
6TH6
 
 | | Cryo-EM Structure of T. kodakarensis 70S ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Matzov, D, Sas-Chen, A, Thomas, J.M, Santangelo, T, Meier, J.L, Schwartz, S, Shalev-Benami, M. | | Deposit date: | 2019-11-18 | | Release date: | 2020-07-29 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Dynamic RNA acetylation revealed by quantitative cross-evolutionary mapping.
Nature, 583, 2020
|
|
5ZCS
 
 | | 4.9 Angstrom Cryo-EM structure of human mTOR complex 2 | | Descriptor: | Rapamycin-insensitive companion of mTOR, Serine/threonine-protein kinase mTOR, Target of rapamycin complex 2 subunit MAPKAP1, ... | | Authors: | Chen, X, Liu, M, Tian, Y, Wang, H, Wang, J, Xu, Y. | | Deposit date: | 2018-02-20 | | Release date: | 2018-03-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Cryo-EM structure of human mTOR complex 2.
Cell Res., 28, 2018
|
|
6NQB
 
 | |
5ZLH
 
 | | Crystal structure of Mn-ProtoporphyrinIX-reconstituted P450BM3 | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, MANGANESE PROTOPORPHYRIN IX | | Authors: | Omura, K, Aiba, Y, Onoda, H, Sugimoto, H, Shoji, O, Watanabe, Y. | | Deposit date: | 2018-03-28 | | Release date: | 2018-08-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Reconstitution of full-length P450BM3 with an artificial metal complex by utilising the transpeptidase Sortase A.
Chem. Commun. (Camb.), 54, 2018
|
|
5YXJ
 
 | | FXR ligand binding domain | | Descriptor: | 2-[benzyl(methyl)amino]ethyl methyl 2,6-dimethyl-4-(3-nitrophenyl)pyridine-3,5-dicarboxylate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Yi, L, Yong, L. | | Deposit date: | 2017-12-05 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | A ligand of drug binding to FXR
To Be Published
|
|
6NER
 
 | | Synthetic Haliangium ochraceum BMC shell | | Descriptor: | BMC-H tandem fusion protein, SULFATE ION | | Authors: | Sutter, M, McGuire, S, Aussignargues, C, Kerfeld, C.A. | | Deposit date: | 2018-12-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.59 Å) | | Cite: | Structural Characterization of a Synthetic Tandem-Domain Bacterial Microcompartment Shell Protein Capable of Forming Icosahedral Shell Assemblies.
ACS Synth Biol, 8, 2019
|
|
6RKE
 
 | | Molybdenum storage protein - P212121, ADP, molybdate | | Descriptor: | (mu3-oxo)-tris(mu2-oxo)-nonakisoxo-trimolybdenum (VI), ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Ermler, U, Bruenle, S. | | Deposit date: | 2019-04-30 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molybdate pumping into the molybdenum storage protein via an ATP-powered piercing mechanism.
Proc.Natl.Acad.Sci.USA, 2019
|
|
6MTD
 
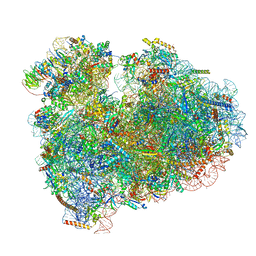 | | Rabbit 80S ribosome with eEF2 and SERBP1 (unrotated state with 40S head swivel) | | Descriptor: | 18S rRNA, 28S rRNA, 5.8S rRNA, ... | | Authors: | Brown, A, Baird, M.R, Yip, M.C.J, Murray, J, Shao, S. | | Deposit date: | 2018-10-19 | | Release date: | 2018-11-21 | | Last modified: | 2019-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structures of translationally inactive mammalian ribosomes.
Elife, 7, 2018
|
|
6N7X
 
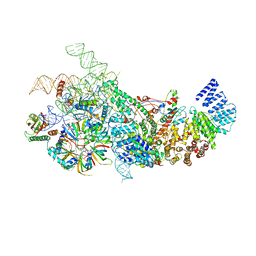 | | S. cerevisiae U1 snRNP | | Descriptor: | 56 kDa U1 small nuclear ribonucleoprotein component, Pre-mRNA-processing factor 39, Protein NAM8, ... | | Authors: | Li, X, Liu, S, Jiang, J, Zhang, L, Espinosa, S, Hill, R.C, Hansen, K.C, Zhou, Z.H, Zhao, R. | | Deposit date: | 2018-11-28 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | CryoEM structure of Saccharomyces cerevisiae U1 snRNP offers insight into alternative splicing.
Nat Commun, 8, 2017
|
|
5OY7
 
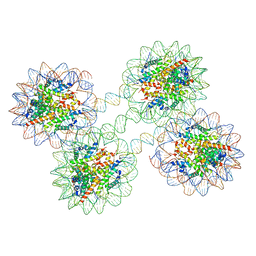 | | Structure of the 4_601_157 tetranucleosome (P1 form) | | Descriptor: | CHLORIDE ION, DNA (619-MER), Histone H2A, ... | | Authors: | Ekundayo, B, Richmond, T.J, Schalch, T. | | Deposit date: | 2017-09-07 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (5.774 Å) | | Cite: | Capturing Structural Heterogeneity in Chromatin Fibers.
J. Mol. Biol., 429, 2017
|
|
6S0Z
 
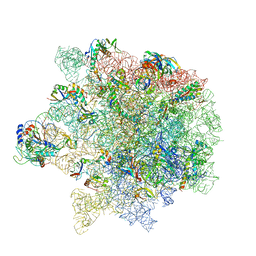 | | Erythromycin Resistant Staphylococcus aureus 50S ribosome (delta R88 A89 uL22) in complex with erythromycin. | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Halfon, Y, Matozv, D, Eyal, Z, Bashan, A, Zimmerman, E, Kjeldgaard, J, Ingmer, H, Yonath, A. | | Deposit date: | 2019-06-18 | | Release date: | 2019-08-21 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Exit tunnel modulation as resistance mechanism of S. aureus erythromycin resistant mutant.
Sci Rep, 9, 2019
|
|
6S05
 
 | |
5YXB
 
 | | A ligand binding to FXR | | Descriptor: | 2-methoxyethyl (2E)-3-phenylprop-2-en-1-yl 2,6-dimethyl-4-(3-nitrophenyl)pyridine-3,5-dicarboxylate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Yi, L, Yong, L. | | Deposit date: | 2017-12-04 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | A ligand binding to FXR
To Be Published
|
|
5YXL
 
 | | A ligand M binding to FXR | | Descriptor: | 2-methoxyethyl propan-2-yl 2,6-dimethyl-4-(3-nitrophenyl)pyridine-3,5-dicarboxylate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Yi, L, Yong, L. | | Deposit date: | 2017-12-05 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | A ligand M binding to FXR
To Be Published
|
|
6NAW
 
 | |
6ND4
 
 | | Conformational switches control early maturation of the eukaryotic small ribosomal subunit | | Descriptor: | 18S rRNA 5' domain start, 5'ETS rRNA, Bud21, ... | | Authors: | Hunziker, M, Barandun, J, Klinge, S. | | Deposit date: | 2018-12-13 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Conformational switches control early maturation of the eukaryotic small ribosomal subunit.
Elife, 8, 2019
|
|
5Z6W
 
 | | Crystal structure of paFAN1 bound to 2nt 5'flap DNA with gap with Manganese | | Descriptor: | DNA (5'-D(P*AP*TP*TP*CP*AP*A)-3'), DNA (5'-D(P*GP*AP*AP*TP*GP*TP*GP*TP*CP*TP*CP*AP*AP*TP*CP*CP*CP*AP*AP*CP*TP*T)-3'), DNA (5'-D(P*GP*TP*TP*GP*GP*GP*AP*TP*TP*G)-3'), ... | | Authors: | Jin, H, Cho, Y. | | Deposit date: | 2018-01-25 | | Release date: | 2018-03-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural mechanism of DNA interstrand cross-link unhooking by the bacterial FAN1 nuclease.
J. Biol. Chem., 293, 2018
|
|
6NF8
 
 | | Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit -Class I | | Descriptor: | 28S ribosomal RNA, mitochondria, 28S ribosomal protein S10, ... | | Authors: | Sharma, M, Koripella, R, Agrawal, R. | | Deposit date: | 2018-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structure of Human Mitochondrial Translation Initiation Factor 3 Bound to the Small Ribosomal Subunit.
iScience, 12, 2019
|
|
6NEQ
 
 | | Structure of human mitochondrial translation initiation factor 3 bound to the small ribosomal subunit-Class-II | | Descriptor: | 28S ribosomal RNA, mitochondrial, 28S ribosomal protein S10, ... | | Authors: | Sharma, M, Koripella, R, Agrawal, R. | | Deposit date: | 2018-12-18 | | Release date: | 2019-02-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.32 Å) | | Cite: | Structure of Human Mitochondrial Translation Initiation Factor 3 Bound to the Small Ribosomal Subunit.
iScience, 12, 2019
|
|
6R8Y
 
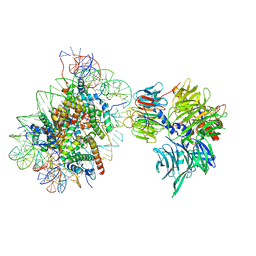 | | Cryo-EM structure of NCP-6-4PP(-1)-UV-DDB | | Descriptor: | DNA damage-binding protein 1, DNA damage-binding protein 2, Histone H2A type 1-B/E, ... | | Authors: | Matsumoto, S, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-04-02 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA damage detection in nucleosomes involves DNA register shifting.
Nature, 571, 2019
|
|
6NHG
 
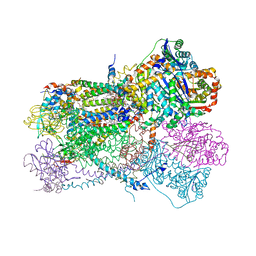 | | Rhodobacter sphaeroides Mitochondrial respiratory chain complex | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Xia, D, Zhou, F, Esser, L. | | Deposit date: | 2018-12-21 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of bacterial cytochromebc1in complex with azoxystrobin reveals a conformational switch of the Rieske iron-sulfur protein subunit.
J.Biol.Chem., 294, 2019
|
|
6TMK
 
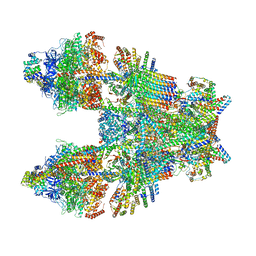 | | Cryo-EM structure of Toxoplasma gondii mitochondrial ATP synthase dimer, composite model | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Dioleoyl-sn-glycero-3-phosphoethanolamine, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Muhleip, A, Kock Flygaard, R, Amunts, A. | | Deposit date: | 2019-12-04 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria.
Nat Commun, 12, 2021
|
|
5ZBO
 
 | | Cryo-EM structure of PCV2 VLPs | | Descriptor: | Capsid protein | | Authors: | Mo, X, Yuan, A.Y. | | Deposit date: | 2018-02-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.12 Å) | | Cite: | Structural roles of PCV2 capsid protein N-terminus in PCV2 particle assembly and identification of PCV2 type-specific neutralizing epitope.
PLoS Pathog., 15, 2019
|
|
