1GII
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE CDK4 INHIBITOR | | Descriptor: | 1-(5-OXO-2,3,5,9B-TETRAHYDRO-1H-PYRROLO[2,1-A]ISOINDOL-9-YL)-3-PYRIDIN-2-YL-UREA, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Ikuta, M, Nishimura, S. | | Deposit date: | 2001-02-06 | | Release date: | 2002-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic approach to identification of cyclin-dependent kinase 4 (CDK4)-specific inhibitors by using CDK4 mimic CDK2 protein.
J.Biol.Chem., 276, 2001
|
|
7UNI
 
 | | De novo designed chlorophyll dimer protein with Zn pheophorbide a methyl ester, SP2-ZnPPaM | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, SP2-ZnPPaM designed chlorophyll dimer protein, ... | | Authors: | Kennedy, M.A, Stoddard, B.L, Ennist, N.M. | | Deposit date: | 2022-04-11 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | De novo design of proteins housing excitonically coupled chlorophyll special pairs.
Nat.Chem.Biol., 2024
|
|
1GIH
 
 | | HUMAN CYCLIN DEPENDENT KINASE 2 COMPLEXED WITH THE CDK4 INHIBITOR | | Descriptor: | 1-(5-OXO-2,3,5,9B-TETRAHYDRO-1H-PYRROLO[2,1-A]ISOINDOL-9-YL)-3-PYRIDIN-2-YL-UREA, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Ikuta, M, Nishimura, S. | | Deposit date: | 2001-02-06 | | Release date: | 2002-02-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystallographic approach to identification of cyclin-dependent kinase 4 (CDK4)-specific inhibitors by using CDK4 mimic CDK2 protein.
J.Biol.Chem., 276, 2001
|
|
7W7E
 
 | | Cryo-EM structure of the alpha2A adrenergic receptor GoA signaling complex bound to a biased agonist | | Descriptor: | 5-(3-bicyclo[4.2.0]octa-1,3,5-trienyl)-1,2,3,6-tetrahydropyridine, Alpha-2A adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Fink, E.A, Shoichet, B.K, Du, Y. | | Deposit date: | 2021-12-04 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based discovery of nonopioid analgesics acting through the alpha 2A -adrenergic receptor.
Science, 377, 2022
|
|
7UZ2
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-fluoro-valienide. | | Descriptor: | (1R,2S,3R,4R)-5-fluoro-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, Beta-galactosidase | | Authors: | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7UZ1
 
 | | Structure of beta-glycosidase from Sulfolobus solfataricus in complex with C5a-bromo-valienide. | | Descriptor: | (1R,2S,3R,4R)-5-bromo-6-(hydroxymethyl)cyclohex-5-ene-1,2,3,4-tetrol, 1,2-ETHANEDIOL, Beta-galactosidase | | Authors: | Danby, P.M, Jeong, A, Sim, L, Sweeney, R.P, Wardman, J.F, Karimi, R, Geissner, A, Worrall, L.J, Strynadka, N.C.J, Withers, S.G. | | Deposit date: | 2022-05-08 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Vinyl Halide-Modified Unsaturated Cyclitols are Mechanism-Based Glycosidase Inhibitors.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
1YLS
 
 | | Crystal structure of selenium-modified Diels-Alder ribozyme complexed with the product of the reaction between N-pentylmaleimide and covalently attached 9-hydroxymethylanthracene | | Descriptor: | (3AS,9AS)-2-PENTYL-4-HYDROXYMETHYL-3A,4,9,9A-TETRAHYDRO-4,9[1',2']-BENZENO-1H-BENZ[F]ISOINDOLE-1,3(2H)-DIONE, MAGNESIUM ION, RNA Diels-Alder ribozyme | | Authors: | Serganov, A, Keiper, S, Malinina, L, Tereshko, V, Skripkin, E, Hobartner, C, Polonskaia, A, Phan, A.T, Wombacher, R, Micura, R, Dauter, Z, Jaschke, A, Patel, D.J. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation.
Nat.Struct.Mol.Biol., 12, 2005
|
|
7XMA
 
 | | Crystal structure of Bovine heart cytochrome c oxidase, apo structure with DMSO | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Takashima, S, Shintani, Y. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7XMB
 
 | | Crystal structure of Bovine heart cytochrome c oxidase, the structure complexed with an allosteric inhibitor T113 | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Nishida, Y, Shinzawa-Itoh, K, Mizuno, N, Kumasaka, T, Yoshikawa, S, Tsukihara, T, Shintani, Y, Takashima, S. | | Deposit date: | 2022-04-25 | | Release date: | 2022-12-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identifying antibiotics based on structural differences in the conserved allostery from mitochondrial heme-copper oxidases.
Nat Commun, 13, 2022
|
|
7WLJ
 
 | | CryoEM structure of human low-voltage activated T-type calcium channel Cav3.3 in complex with mibefradil (MIB) | | Descriptor: | (1S,2S)-2-(2-{[3-(1H-benzimidazol-2-yl)propyl](methyl)amino}ethyl)-6-fluoro-1-(propan-2-yl)-1,2,3,4-tetrahydronaphthalen-2-yl methoxyacetate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | He, L, Yu, Z, Dong, Y, Chen, Q, Zhao, Y. | | Deposit date: | 2022-01-13 | | Release date: | 2022-05-04 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure, gating, and pharmacology of human Ca V 3.3 channel.
Nat Commun, 13, 2022
|
|
1HBV
 
 | |
7W9P
 
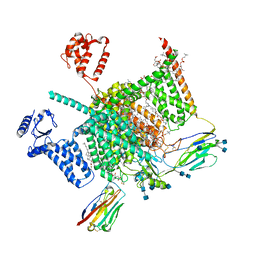 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (S6IV pi helix conformer) | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P, Shen, H. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7W9T
 
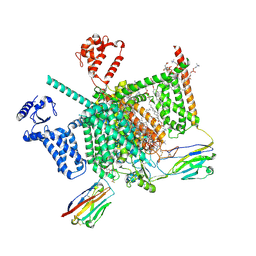 | | Cryo-EM structure of human Nav1.7(E406K) in complex with auxiliary beta subunits, huwentoxin-IV and saxitoxin (S6IV alpha helix conformer) | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Yan, N, Huang, G, Liu, D, Wei, P. | | Deposit date: | 2021-12-10 | | Release date: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | High-resolution structures of human Na v 1.7 reveal gating modulation through alpha-pi helical transition of S6 IV.
Cell Rep, 39, 2022
|
|
7WR6
 
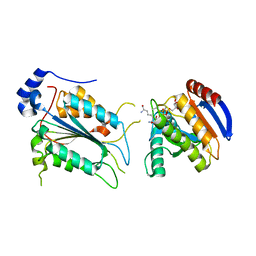 | | Crystal structure of ADP-riboxanated caspase-4 in complex with Af1521 | | Descriptor: | ADP-ribose glycohydrolase AF_1521, Caspase-4, [[(3~{a}~{S},5~{R},6~{R},6~{a}~{R})-2-azanylidene-3-[(4~{R})-4-azanyl-5-oxidanylidene-pentyl]-6-oxidanyl-3~{a},5,6,6~{a}-tetrahydrofuro[2,3-d][1,3]oxazol-5-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Hou, Y.J, Zeng, H, Shao, F, Ding, J. | | Deposit date: | 2022-01-26 | | Release date: | 2023-01-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural mechanisms of calmodulin activation of Shigella effector OspC3 to ADP-riboxanate caspase-4/11 and block pyroptosis.
Nat.Struct.Mol.Biol., 30, 2023
|
|
2GKU
 
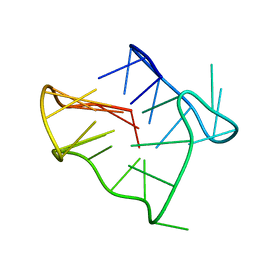 | | Monomeric human telomere DNA tetraplex with 3+1 strand fold topology, two edgewise loops and double-chain reversal loop, NMR, 12 structures | | Descriptor: | Human telomere DNA | | Authors: | Luu, K.N, Phan, A.T, Kuryavyi, V.V, Lacroix, L, Patel, D.J. | | Deposit date: | 2006-04-03 | | Release date: | 2006-08-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the human telomere in k(+) solution: an intramolecular (3 + 1) g-quadruplex scaffold.
J.Am.Chem.Soc., 128, 2006
|
|
7XBO
 
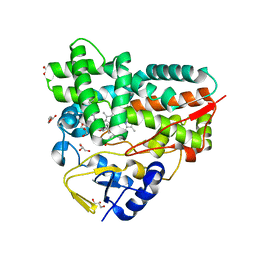 | | Crystal Structure of 10-dml-bound cytochrome P450 PikC with the unnatural amino acid p-Acetyl-L-Phenylalanine incorporated at position 238 | | Descriptor: | (3R,4S,5S,7R,9E,11R,12R)-12-ETHYL-4-HYDROXY-3,5,7,11-TETRAMETHYLOXACYCLODODEC-9-ENE-2,8-DIONE, Cytochrome P450 monooxygenase PikC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Li, G.B, Pan, Y.J, Li, S.Y, Gao, X. | | Deposit date: | 2022-03-21 | | Release date: | 2023-02-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | New mechanistic insight of cytochrome P450 PikC gained from site-specific mutagenesis by non-coding amino acids
Nat Commun, 2023
|
|
7XP7
 
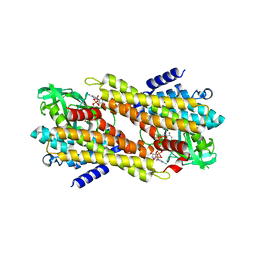 | | Crystal Structure of the Flavoprotein ColB1 Catalyzing Assembly Line-Tethered Cysteine Dehydrogenation | | Descriptor: | Cyclohexanecarboxyl-CoA dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Ma, X.Y, Tang, Z.J, Liu, W, Ma, M. | | Deposit date: | 2022-05-03 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Mechanistic Insights into ColB1, a Flavoprotein Functioning in-trans in the 2,2'-Bipyridine Assembly Line for Cysteine Dehydrogenation.
Acs Chem.Biol., 18, 2023
|
|
7WZ9
 
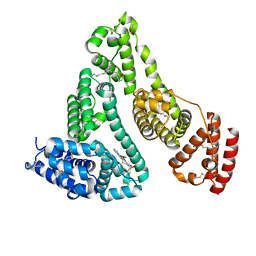 | | HSA-In agent complex | | Descriptor: | 16-chloranyl-~{N},~{N}-dimethyl-15-thia-1$l^{4},12$l^{4},13-triaza-16$l^{4}-indatetracyclo[8.6.0.0^{2,7}.0^{12,16}]hexadeca-1,3,5,7,9,11,13-heptaen-14-amine, PALMITIC ACID, Serum albumin | | Authors: | Zhang, Z.L, Yang, F. | | Deposit date: | 2022-02-17 | | Release date: | 2023-02-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structure of HSA-In agent complex
To Be Published
|
|
2BVW
 
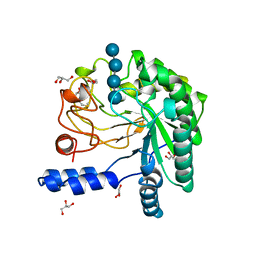 | | CELLOBIOHYDROLASE II (CEL6A) FROM HUMICOLA INSOLENS IN COMPLEX WITH GLUCOSE AND CELLOTETRAOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE II, GLYCEROL, ... | | Authors: | Varrot, A, Davies, G.J, Schulein, M. | | Deposit date: | 1999-02-18 | | Release date: | 2000-02-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural changes of the active site tunnel of Humicola insolens cellobiohydrolase, Cel6A, upon oligosaccharide binding.
Biochemistry, 38, 1999
|
|
7X4U
 
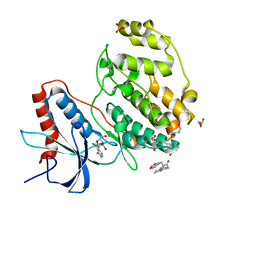 | | Crystal structure of ERK2 with an allosteric inhibitor 2 | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, ... | | Authors: | Yoshida, M, Kinoshita, T. | | Deposit date: | 2022-03-03 | | Release date: | 2023-03-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for producing allosteric ERK2 inhibitors
To Be Published
|
|
1Y3P
 
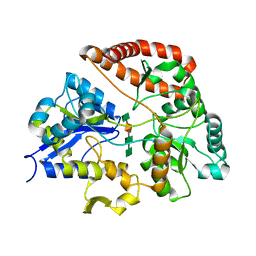 | | Structure of AlgQ1, alginate-binding protein, complexed with an alginate tetrasaccharide | | Descriptor: | AlgQ1, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-D-mannopyranuronic acid | | Authors: | Momma, K, Mishima, Y, Hashimoto, W, Mikami, B, Murata, K. | | Deposit date: | 2004-11-26 | | Release date: | 2005-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Direct Evidence for Sphingomonas sp. A1 Periplasmic Proteins as Macromolecule-Binding Proteins Associated with the ABC Transporter: Molecular Insights into Alginate Transport in the Periplasm(,)
Biochemistry, 44, 2005
|
|
2CGF
 
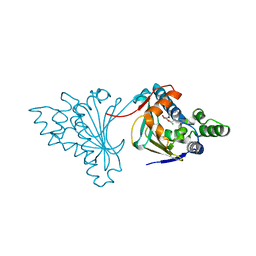 | | A RADICICOL ANALOGUE BOUND TO THE ATP BINDING SITE OF THE N-TERMINAL DOMAIN OF THE YEAST HSP90 CHAPERONE | | Descriptor: | (5Z)-13-CHLORO-14,16-DIHYDROXY-3,4,7,8,9,10-HEXAHYDRO-1H-2-BENZOXACYCLOTETRADECINE-1,11(12H)-DIONE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H, Moody, C.J. | | Deposit date: | 2006-03-02 | | Release date: | 2006-11-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of Hsp90 with Synthetic Macrolactones: Synthesis and Structural and Biological Evaluation of Ring and Conformational Analogs of Radicicol.
Chem.Biol., 13, 2006
|
|
7WXP
 
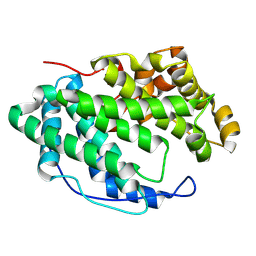 | |
2BD7
 
 | | Porcine pancreatic elastase complexed with beta-casomorphin-7 and Arg-Phe at pH 5.0 (50 min soak) | | Descriptor: | ARGININE, CALCIUM ION, Elastase-1, ... | | Authors: | Liu, B, Schofield, C.J, Wilmouth, R.C. | | Deposit date: | 2005-10-20 | | Release date: | 2006-05-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural analyses on intermediates in serine protease catalysis
J.Biol.Chem., 281, 2006
|
|
1H6H
 
 | | Structure of the PX domain from p40phox bound to phosphatidylinositol 3-phosphate | | Descriptor: | 2-(BUTANOYLOXY)-1-{[(HYDROXY{[2,3,4,6-TETRAHYDROXY-5-(PHOSPHONOOXY)CYCLOHEXYL]OXY}PHOSPHORYL)OXY]METHYL}ETHYL BUTANOATE, GLYCEROL, NEUTROPHIL CYTOSOL FACTOR 4 | | Authors: | Karathanassis, D, Bravo, J, Pacold, M, Perisic, O, Williams, R.L. | | Deposit date: | 2001-06-15 | | Release date: | 2001-11-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of the Px Domain from P40Phox Bound to Phosphatidylinositol 3-Phosphate
Mol.Cell, 8, 2001
|
|
