5OJL
 
 | | Imine Reductase from Aspergillus terreus in complex with NADPH4 and dibenz[c,e]azepine | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, 5-methyl-7~{H}-benzo[d][2]benzazepine, Imine reductase | | Authors: | Sharma, M, Grogan, G. | | Deposit date: | 2017-07-21 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Biocatalytic Routes to Enantiomerically Enriched Dibenz[c,e]azepines.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2JBP
 
 | | Protein kinase MK2 in complex with an inhibitor (crystal form-2, co- crystallization) | | Descriptor: | 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE, MAP KINASE-ACTIVATED PROTEIN KINASE 2 | | Authors: | Hillig, R.C, Eberspaecher, U, Monteclaro, F, Huber, M, Nguyen, D, Mengel, A, Muller-Tiemann, B, Egner, U. | | Deposit date: | 2006-12-09 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis for a high affinity inhibitor bound to protein kinase MK2.
J. Mol. Biol., 369, 2007
|
|
2ZBD
 
 | | Crystal Structure of the SR Calcium Pump with Bound Aluminium Fluoride, ADP and Calcium | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Toyoshima, C, Nomura, H, Tsuda, T, Ogawa, H, Norimatsu, Y. | | Deposit date: | 2007-10-20 | | Release date: | 2007-11-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues
Nature, 432, 2004
|
|
3VDI
 
 | | Structure of the FMO protein from Pelodictyon phaeum | | Descriptor: | BACTERIOCHLOROPHYLL A, TETRAETHYLENE GLYCOL, bacteriochlorophyll A protein | | Authors: | Tronrud, D.E, Larson, C.R, Seng, C.O, Lauman, L, Matthies, H.J, Wen, J, Blankenship, R.E, Allen, J.P. | | Deposit date: | 2012-01-05 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Reinterpretation of the electron density at the site of the eighth bacteriochlorophyll in the FMO protein from Pelodictyon phaeum.
Photosynth.Res., 112, 2012
|
|
1EKI
 
 | | AVERAGE SOLUTION STRUCTURE OF D(TTGGCCAA)2 BOUND TO CHROMOMYCIN-A3 AND COBALT | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-4-O-acetyl-2,6-dideoxy-beta-D-galactopyranose, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-beta-D-Olivopyranose-(1-3)-beta-D-Olivopyranose, ... | | Authors: | Gochin, M. | | Deposit date: | 2000-03-08 | | Release date: | 2000-03-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A high-resolution structure of a DNA-chromomycin-Co(II) complex determined from pseudocontact shifts in nuclear magnetic resonance.
Structure Fold.Des., 8, 2000
|
|
3VM6
 
 | | Crystal structure of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1 in complex with alpha-D-ribose-1,5-bisphosphate | | Descriptor: | 1,5-di-O-phosphono-alpha-D-ribofuranose, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nakamura, A, Fujihashi, M, Aono, R, Sato, T, Nishiba, Y, Yoshida, S, Yano, A, Atomi, H, Imanaka, T, Miki, K. | | Deposit date: | 2011-12-08 | | Release date: | 2012-04-25 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Dynamic, ligand-dependent conformational change triggers reaction of ribose-1,5-bisphosphate isomerase from Thermococcus kodakarensis KOD1
J.Biol.Chem., 287, 2012
|
|
1E80
 
 | | Endothiapepsin complex with renin inhibitor MERCK-KGAA-EMD56133 | | Descriptor: | 4-amino-N-{(1R,8R,9R,13R)-16-(4-amino-2-methylpyrimidin-5-yl)-1-benzyl-8-(cyclohexylmethyl)-9-hydroxy-13-[(1S)-1-methylpropyl]-2,6,11,14-tetraoxo-3,7,12,15-tetraazahexadec-1-yl}piperidine-1-carboxamide, ENDOTHIAPEPSIN | | Authors: | Read, J.A, Cooper, J.B, Toldo, L, Rippmann, F, Raddatz, P. | | Deposit date: | 2000-09-15 | | Release date: | 2000-09-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Refinement of Four Endothiapepsin Inhibitor Complexes. Crystallographic Studies of Cytochrome Ch from Methylobacterium Extorquens and Inhibitor Complexes of Aspartic Proteinases.
Ph D Thesis, 1999
|
|
1ZKG
 
 | |
2D82
 
 | | Target Structure-Based Discovery of Small Molecules that Block Human p53 and CREB Binding Protein (CBP) Association | | Descriptor: | 9-ACETYL-2,3,4,9-TETRAHYDRO-1H-CARBAZOL-1-ONE, CREB-binding protein | | Authors: | Sachchidanand, Resnick-Silverman, L, Yan, S, Mujtaba, S, Liu, W.J, Zeng, L, Manfredi, J.J, Zhou, M.M. | | Deposit date: | 2005-12-01 | | Release date: | 2006-04-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Target structure-based discovery of small molecules that block human p53 and CREB binding protein association
Chem.Biol., 13, 2006
|
|
1EXA
 
 | | ENANTIOMER DISCRIMINATION ILLUSTRATED BY CRYSTAL STRUCTURES OF THE HUMAN RETINOIC ACID RECEPTOR HRARGAMMA LIGAND BINDING DOMAIN: THE COMPLEX WITH THE ACTIVE R-ENANTIOMER BMS270394. | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, R-3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, RETINOIC ACID RECEPTOR GAMMA-2 | | Authors: | Klaholz, B.P, Mitschler, A, Belema, M, Zusi, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-05-02 | | Release date: | 2000-06-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARgamma.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1JP3
 
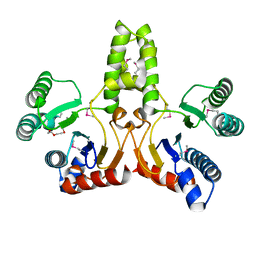 | | Structure of E.coli undecaprenyl pyrophosphate synthase | | Descriptor: | 2-(2-{2-[2-(2-{2-[2-(2-{2-[4-(1,1,3,3-TETRAMETHYL-BUTYL)-PHENOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, undecaprenyl pyrophosphate synthase | | Authors: | Ko, T.P, Chen, Y.K, Robinson, H, Tsai, P.C, Gao, Y.G, Chen, A.P.C, Wang, A.H.J, Liang, P.H. | | Deposit date: | 2001-07-31 | | Release date: | 2001-08-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of product chain length determination and the role of a flexible loop in Escherichia coli undecaprenyl-pyrophosphate synthase catalysis.
J.Biol.Chem., 276, 2001
|
|
5P9F
 
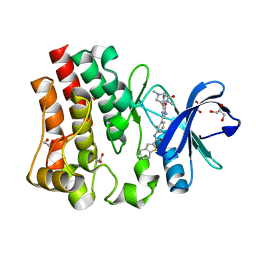 | | BTK IN COMPLEX WITH GDC-0834 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-{3-[6-({4-[(2R)-1,4-dimethyl-3-oxopiperazin-2-yl]phenyl}amino)-4-methyl-5-oxo-4,5-dihydropyrazin-2-yl]-2-methylphenyl }-4,5,6,7-tetrahydro-1-benzothiophene-2-carboxamide, ... | | Authors: | Gardberg, A.S. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Ability of Bruton's Tyrosine Kinase Inhibitors to Sequester Y551 and Prevent Phosphorylation Determines Potency for Inhibition of Fc Receptor but not B-Cell Receptor Signaling.
Mol. Pharmacol., 91, 2017
|
|
1E61
 
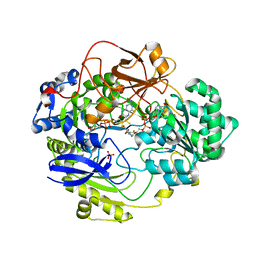 | | OXIDIZED DMSO REDUCTASE EXPOSED TO HEPES - Structure II BUFFER | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Bailey, S, Bennett, B, Adams, B, Smith, A.T, Bray, R.C. | | Deposit date: | 2000-08-06 | | Release date: | 2000-08-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible Dissociation of Thiolate Ligands from Molybdenum in an Enzyme of the Dimethyl Sulfoxide Reductase Family
Biochemistry, 39, 2000
|
|
1EXX
 
 | | ENANTIOMER DISCRIMINATION ILLUSTRATED BY CRYSTAL STRUCTURES OF THE HUMAN RETINOIC ACID RECEPTOR HRARGAMMA LIGAND BINDING DOMAIN: THE COMPLEX WITH THE INACTIVE S-ENANTIOMER BMS270395. | | Descriptor: | 3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, DODECYL-ALPHA-D-MALTOSIDE, RETINOIC ACID RECEPTOR GAMMA-2 | | Authors: | Klaholz, B.P, Mitschler, A, Belema, M, Zusi, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-05-05 | | Release date: | 2000-06-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARgamma.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E60
 
 | | OXIDIZED DMSO REDUCTASE EXPOSED TO HEPES - Structure II BUFFER | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, Dimethyl sulfoxide/trimethylamine N-oxide reductase, MOLYBDENUM (IV)OXIDE, ... | | Authors: | Bailey, S, Bennett, B, Adams, B, Smith, A.T, Bray, R.C. | | Deposit date: | 2000-08-06 | | Release date: | 2000-08-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Reversible Dissociation of Thiolate Ligands from Molybdenum in an Enzyme of the Dimethyl Sulfoxide Reductase Family
Biochemistry, 39, 2000
|
|
2ZJH
 
 | |
5OST
 
 | | Beta-glucosidase from Thermoanaerobacterium xylolyticum GH116 in complex with Gluco-1H-imidazole | | Descriptor: | (4~{S},5~{S},6~{R},7~{R})-7-(hydroxymethyl)-4,5,6,7-tetrahydro-1~{H}-benzimidazole-4,5,6-triol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Davies, G.J, Offen, W.A. | | Deposit date: | 2017-08-18 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gluco-1 H-imidazole: A New Class of Azole-Type beta-Glucosidase Inhibitor.
J. Am. Chem. Soc., 140, 2018
|
|
3VRC
 
 | | Crystal structure of cytochrome c' from Thermochromatium tepidum | | Descriptor: | CADMIUM ION, CHLORIDE ION, Cytochrome c', ... | | Authors: | Hirano, Y, Kimura, Y, Suzuki, H, Miki, K, Wang, Z.-Y. | | Deposit date: | 2012-04-09 | | Release date: | 2012-09-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure Analysis and Comparative Characterization of the Cytochrome c' and Flavocytochrome c from Thermophilic Purple Photosynthetic Bacterium Thermochromatium tepidum
Biochemistry, 51, 2012
|
|
2N2K
 
 | | Ensemble structure of the closed state of Lys63-linked diubiquitin in the absence of a ligand | | Descriptor: | S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, ubiquitin | | Authors: | Liu, Z, Gong, Z, Tang, C. | | Deposit date: | 2015-05-10 | | Release date: | 2015-07-08 | | Last modified: | 2015-09-23 | | Method: | SOLUTION NMR | | Cite: | Lys63-linked ubiquitin chain adopts multiple conformational states for specific target recognition.
Elife, 4, 2015
|
|
5ONO
 
 | | Crystal Structure of Ectoine Synthase from P. lautus | | Descriptor: | (4S)-2-METHYL-1,4,5,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, FE (III) ION, L-ectoine synthase | | Authors: | Bremer, E. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Illuminating the catalytic core of ectoine synthase through structural and biochemical analysis.
Sci Rep, 9, 2019
|
|
8QQJ
 
 | | CryoEM structure of the type IV pilin PilA5 from Thermus thermophilus | | Descriptor: | 7-Acetamido-5-acetimidoyl-3,5,7,9-tetradeoxy-L-glycero-L-manno-nonulosonic aci-(1-4)-alpha-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose, MAGNESIUM ION, Type IV narrow pilus major component PilA5 | | Authors: | Gold, V.A.M, Neuhaus, A, Gaines, M, Isupov, M, McLaren, M. | | Deposit date: | 2023-10-04 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | CryoEM structure of the type IV pilin PilA4 from Thermus thermophilus
To Be Published
|
|
5OX5
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with CCT6, a GSK1278863-related compound | | Descriptor: | (6-hydroxy-1,3-dimethyl-2,4-dioxo-1,2,3,4-tetrahydropyrimidine-5-carbonyl)glycine, BICARBONATE ION, Egl nine homolog 1, ... | | Authors: | Chowdhury, R, Thinnes, C.C, Schofield, C.J. | | Deposit date: | 2017-09-06 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Molecular and cellular mechanisms of HIF prolyl hydroxylase inhibitors in clinical trials.
Chem Sci, 8, 2017
|
|
5OY0
 
 | | Structure of synechocystis photosystem I trimer at 2.5A resolution | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (3'R)-3'-hydroxy-beta,beta-caroten-4-one, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, ... | | Authors: | Nelson, N, Malavath, T, Caspy, I. | | Deposit date: | 2017-09-07 | | Release date: | 2018-02-21 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Structure and function of wild-type and subunit-depleted photosystem I in Synechocystis.
Biochim. Biophys. Acta, 1859, 2018
|
|
2ZJY
 
 | | Structure of the K349P mutant of Gi alpha 1 subunit bound to ALF4 and GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i), alpha-1 subunit, ... | | Authors: | Morikawa, T, Muroya, A, Sugio, S, Wakamatsu, K, Kohno, T. | | Deposit date: | 2008-03-11 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | How GPCRs activate G proteins: Structural changes form C-terminal tail to GDP binding pocket
To be Published
|
|
3VDX
 
 | |
