5XF1
 
 | | Structure of the Full-length glucagon class B G protein-coupled receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, H, Qiao, A, Yang, D, Yang, L, Dai, A, de Graaf, C, Reedtz-Runge, S, Dharmarajan, V, Zhang, H, Han, G.W, Grant, T, Sierra, R, Weierstall, U, Nelson, G, Liu, W, Wu, Y, Ma, L, Cai, X, Lin, G, Wu, X, Geng, Z, Dong, Y, Song, G, Griffin, P, Lau, J, Cherezov, V, Yang, H, Hanson, M, Stevens, R, Jiang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structure of the full-length glucagon class B G-protein-coupled receptor.
Nature, 546, 2017
|
|
6RT5
 
 | |
3PL7
 
 | |
3UUS
 
 | |
8F6H
 
 | | Cryo-EM structure of a Zinc-loaded asymmetrical TMD D70A mutant of the YiiP-Fab complex | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab2r heavy chain, Fab2r light chain, ... | | Authors: | Lopez-Redondo, M.L, Hussein, A.K, Stokes, D.L. | | Deposit date: | 2022-11-16 | | Release date: | 2023-02-08 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Energy coupling and stoichiometry of Zn 2+ /H + antiport by the prokaryotic cation diffusion facilitator YiiP.
Elife, 12, 2023
|
|
6RSW
 
 | | HFD domain of mouse CAP1 bound to the pointed end of G-actin | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, Actin, ... | | Authors: | Kotila, T, Kogan, K, Lappalainen, P. | | Deposit date: | 2019-05-22 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Mechanism of synergistic actin filament pointed end depolymerization by cyclase-associated protein and cofilin.
Nat Commun, 10, 2019
|
|
6LLZ
 
 | |
3UZY
 
 | | Crystal structure of 5beta-reductase (AKR1D1) E120H mutant in complex with NADP+ and 5beta-dihydrotestosterone | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, 5-beta-DIHYDROTESTOSTERONE, CHLORIDE ION, ... | | Authors: | Chen, M, Christianson, D.W, Penning, T.M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | Conversion of Human Steroid 5beta-Reductase (AKR1D1) into 3β-Hydroxysteroid Dehydrogenase by Single Point Mutation E120H: EXAMPLE OF PERFECT ENZYME ENGINEERING.
J.Biol.Chem., 287, 2012
|
|
3GKW
 
 | | Crystal structure of the Fab fragment of Nimotuzumab. An anti-epidermal growth factor receptor antibody | | Descriptor: | DI(HYDROXYETHYL)ETHER, Heavy chain of the antibody Nimotuzumab, Light chain of the antibody Nimotuzumab, ... | | Authors: | Talavera, A, Friemann, R, Martinez-Fleites, C, Moreno, E, Krengel, U. | | Deposit date: | 2009-03-11 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nimotuzumab, an antitumor antibody that targets the epidermal growth factor receptor, blocks ligand binding while permitting the active receptor conformation
Cancer Res., 69, 2009
|
|
6RVV
 
 | | Structure of left-handed protein cage consisting of 24 eleven-membered ring proteins held together by gold (I) bridges. | | Descriptor: | GOLD ION, Transcription attenuation protein MtrB | | Authors: | Malay, A.D, Miyazaki, N, Biela, A.P, Iwasaki, K, Heddle, J.G. | | Deposit date: | 2019-06-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ultra-stable gold-coordinated protein cage displaying reversible assembly.
Nature, 569, 2019
|
|
3Q35
 
 | | Structure of the Rtt109-AcCoA/Vps75 complex and implications for chaperone-mediated histone acetylation | | Descriptor: | 1,2-ETHANEDIOL, ACETYL COENZYME *A, Histone acetyltransferase, ... | | Authors: | Tang, Y, Yuan, H, Meeth, K, Marmorstein, R. | | Deposit date: | 2010-12-21 | | Release date: | 2011-02-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the Rtt109-AcCoA/Vps75 Complex and Implications for Chaperone-Mediated Histone Acetylation.
Structure, 19, 2011
|
|
6RV7
 
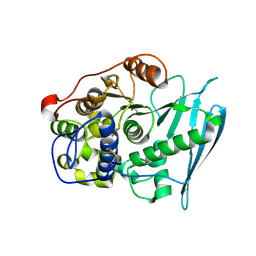 | | Crystal Structure of Glucuronoyl Esterase from Cerrena unicolor inactive S270A variant in complex with the aldouronic acid UXXR | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-Xylitol, ... | | Authors: | Ernst, H.A, Mosbech, C, Langkilde, A, Westh, P, Meyer, A, Agger, J.W, Larsen, S. | | Deposit date: | 2019-05-31 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | The structural basis of fungal glucuronoyl esterase activity on natural substrates.
Nat Commun, 11, 2020
|
|
8EM7
 
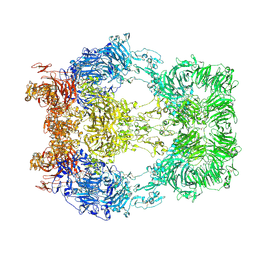 | | Cryo-EM structure of LRP2 at pH 5.2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Beenken, A, Cerutti, G, Fitzpatrick, A.W, Barasch, J, Shapiro, L. | | Deposit date: | 2022-09-27 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of LRP2 reveal a molecular machine for endocytosis.
Cell, 186, 2023
|
|
3GLY
 
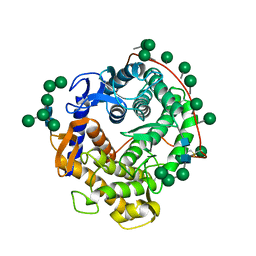 | | REFINED CRYSTAL STRUCTURES OF GLUCOAMYLASE FROM ASPERGILLUS AWAMORI VAR. X100 | | Descriptor: | GLUCOAMYLASE-471, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Aleshin, A.E, Hoffman, C, Firsov, L.M, Honzatko, R.B. | | Deposit date: | 1994-06-03 | | Release date: | 1994-11-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined crystal structures of glucoamylase from Aspergillus awamori var. X100.
J.Mol.Biol., 238, 1994
|
|
4BPJ
 
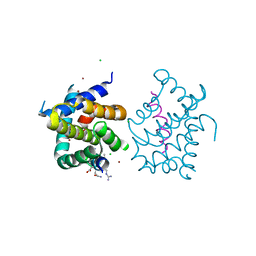 | | Mcl-1 bound to alpha beta Puma BH3 peptide 3 | | Descriptor: | ALPHA BETA BH3-PEPTIDE, CHLORIDE ION, FUSION PROTEIN CONSISTING OF INDUCED MYELOID LEUKEMIA CELL DIFFERENTIATION PROTEIN MCL-1 HOMOLOG, ... | | Authors: | Smith, B.J, Lee, E.F, Checco, J.W, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2013-05-27 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Structure-Guided Rational Design of Alpha/Beta-Peptide Foldamers with High Affinity for Bcl-2 Family Prosurvival Proteins.
Chembiochem, 14, 2013
|
|
4BPI
 
 | | Mcl-1 bound to alpha beta Puma BH3 peptide 2 | | Descriptor: | ALPHA BETA BH3PEPTIDE, CADMIUM ION, FUSION PROTEIN CONSISTING OF INDUCED MYELOID LEUKEMIA CELL DIFFERENTIATION PROTEIN MCL-1 HOMOLOG | | Authors: | Smith, B.J, Lee, E.F, Checco, J.W, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2013-05-27 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.982 Å) | | Cite: | Structure-Guided Rational Design of Alpha/Beta-Peptide Foldamers with High Affinity for Bcl-2 Family Prosurvival Proteins.
Chembiochem, 14, 2013
|
|
3LUM
 
 | | Structure of ulilysin mutant M290L | | Descriptor: | ARGININE, CALCIUM ION, GLYCEROL, ... | | Authors: | Tallant, C, Garcia-Castellanos, R, Baumann, U, Gomis-Ruth, F.X. | | Deposit date: | 2010-02-18 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | On the relevance of the Met-turn methionine in metzincins.
J.Biol.Chem., 285, 2010
|
|
3V1N
 
 | | Crystal Structure of the H265Q mutant of a C-C hydrolase, BphD from Burkholderia xenovorans LB400, after exposure to its substrate HOPDA | | Descriptor: | (3E)-2,6-DIOXO-6-PHENYLHEX-3-ENOATE, 2-hydroxy-6-oxo-6-phenylhexa-2,4-dienoate hydrolase, BENZOIC ACID, ... | | Authors: | Ghosh, S, Bolin, J.T. | | Deposit date: | 2011-12-09 | | Release date: | 2012-03-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Identification of an Acyl-Enzyme Intermediate in a meta-Cleavage Product Hydrolase Reveals the Versatility of the Catalytic Triad.
J.Am.Chem.Soc., 134, 2012
|
|
3LMU
 
 | | Crystal structure of DTD from Plasmodium falciparum | | Descriptor: | D-tyrosyl-tRNA(Tyr) deacylase, IODIDE ION | | Authors: | Manickam, Y, Bhatt, T.K, Khan, S, Sharma, A. | | Deposit date: | 2010-02-01 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of D-tyrosyl-tRNATyr deacylase using home-source Cu Kalpha and moderate-quality iodide-SAD data: structural polymorphism and HEPES-bound enzyme states
Acta Crystallogr.,Sect.D, 66, 2010
|
|
6M17
 
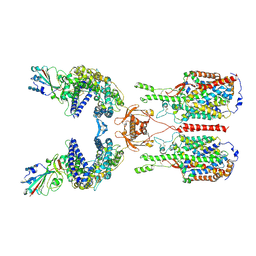 | | The 2019-nCoV RBD/ACE2-B0AT1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, R.H, Zhang, Y.Y, Li, Y.N, Xia, L, Guo, Y.Y, Zhou, Q. | | Deposit date: | 2020-02-24 | | Release date: | 2020-03-11 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2.
Science, 367, 2020
|
|
4BPK
 
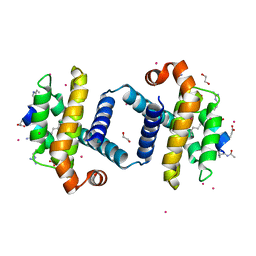 | | Bcl-xL bound to alpha beta Puma BH3 peptide 5 | | Descriptor: | 1,2-ETHANEDIOL, ALPHA BETA BH3-PEPTIDE, BCL-2-LIKE PROTEIN 1, ... | | Authors: | Smith, B.J, Lee, E.F, Checco, J.W, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2013-05-27 | | Release date: | 2014-04-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.756 Å) | | Cite: | Structure-Guided Rational Design of Alpha/Beta-Peptide Foldamers with High Affinity for Bcl-2 Family Prosurvival Proteins.
Chembiochem, 14, 2013
|
|
8EM4
 
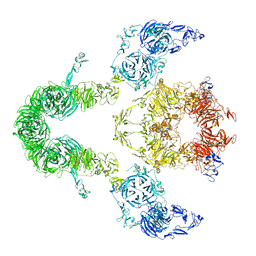 | | Cryo-EM structure of LRP2 at pH 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Beenken, A, Cerutti, G, Brasch, J, Fitzpatrick, A.W, Barasch, J, Shapiro, L. | | Deposit date: | 2022-09-26 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-08 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structures of LRP2 reveal a molecular machine for endocytosis.
Cell, 186, 2023
|
|
8F6E
 
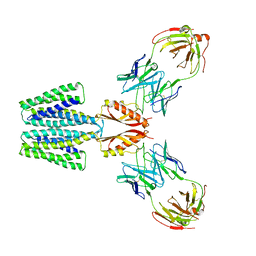 | | Cryo-EM structure of a Zinc-loaded wild-type YiiP-Fab complex | | Descriptor: | Cadmium and zinc efflux pump FieF, Fab heavy chain, Fab light chain, ... | | Authors: | Lopez-Redondo, M.L, Hussein, A.K, Stokes, D.L. | | Deposit date: | 2022-11-16 | | Release date: | 2023-02-08 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Energy coupling and stoichiometry of Zn 2+ /H + antiport by the prokaryotic cation diffusion facilitator YiiP.
Elife, 12, 2023
|
|
6RZ7
 
 | | Crystal structure of the human cysteinyl leukotriene receptor 2 in complex with ONO-2570366 (F222 space group) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{S})-8-[[4-[4-(2-chloranyl-5-fluoranyl-phenyl)butoxy]phenyl]carbonylamino]-4-(4-oxidanyl-4-oxidanylidene-butyl)-2,3- dihydro-1,4-benzoxazine-2-carboxylic acid, CHOLESTEROL, ... | | Authors: | Gusach, A, Luginina, A, Marin, E, Brouillette, R.L, Besserer-Offroy, E, Longpre, J.M, Ishchenko, A, Popov, P, Fujimoto, T, Maruyama, T, Stauch, B, Ergasheva, M, Romanovskaya, D, Stepko, A, Kovalev, K, Shevtsov, M, Gordeliy, V, Han, G.W, Sarret, P, Katritch, V, Borshchevskiy, V, Mishin, A, Cherezov, V. | | Deposit date: | 2019-06-12 | | Release date: | 2019-12-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis of ligand selectivity and disease mutations in cysteinyl leukotriene receptors.
Nat Commun, 10, 2019
|
|
3H90
 
 | |
