6FOF
 
 | | Crystal structure of a crystallized variant of h-Gal3: Gal-3[NTS/VII-IX] | | Descriptor: | Galectin-3,Galectin-3, SULFATE ION, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Romero, A, Flores-Ibarra, A, Medrano, F.J. | | Deposit date: | 2018-02-07 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallization of a human galectin-3 variant with two ordered segments in the shortened N-terminal tail.
Sci Rep, 8, 2018
|
|
6GC8
 
 | | 50S ribosomal subunit assembly intermediate - 50S rec* | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Nikolay, R, Hilal, T, Qin, B, Loerke, J, Buerger, J, Mielke, T, Spahn, C.M.T. | | Deposit date: | 2018-04-17 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural Visualization of the Formation and Activation of the 50S Ribosomal Subunit during In Vitro Reconstitution.
Mol. Cell, 70, 2018
|
|
4KCL
 
 | | Structure of neuronal nitric oxide synthase heme domain in complex with N-(4-(2-((3-(thiophene-2-carboximidamido)benzyl)amino)ethyl)phenyl)thiophene-2-carboximidamide | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-(4-{2-[(3-{[(E)-imino(thiophen-2-yl)methyl]amino}benzyl)amino]ethyl}phenyl)thiophene-2-carboximidamide, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Potent and Selective Double-Headed Thiophene-2-carboximidamide Inhibitors of Neuronal Nitric Oxide Synthase for the Treatment of Melanoma.
J.Med.Chem., 57, 2014
|
|
6FS2
 
 | | MCL1 in complex with indole acid ligand | | Descriptor: | 7-(2-methylphenyl)-3-[3-(5,6,7,8-tetrahydronaphthalen-1-yloxy)propyl]-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Hargreaves, D. | | Deposit date: | 2018-02-18 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia.
Nat Commun, 9, 2018
|
|
4KCS
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with N-(3-((ethyl(3-fluorophenethyl)amino)methyl)phenyl)thiophene-2-carboximidamide | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, GLYCEROL, ... | | Authors: | Chreifi, G, Li, H, Poulos, T.L. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Potent and Selective Double-Headed Thiophene-2-carboximidamide Inhibitors of Neuronal Nitric Oxide Synthase for the Treatment of Melanoma.
J.Med.Chem., 57, 2014
|
|
3QBG
 
 | | Anion-free blue form of pharaonis halorhodopsin | | Descriptor: | BACTERIORUBERIN, Halorhodopsin, RETINAL, ... | | Authors: | Kouyama, T, Kanada, S. | | Deposit date: | 2011-01-13 | | Release date: | 2011-08-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of an O-like blue form and an anion-free yellow form of pharaonis halorhodopsin
J.Mol.Biol., 413, 2011
|
|
6FSX
 
 | | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography | | Descriptor: | Fluoroacetate dehalogenase | | Authors: | Schulz, E.C, Mehrabi, P, Mueller-werkmeiser, H, Tellkamp, F, Jha, A, Stuart, W, Persch, E, De Gasparo, R, Diederich, F, Pai, E, Miller, D. | | Deposit date: | 2018-02-20 | | Release date: | 2018-10-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The hit-and-return system enables efficient time-resolved serial synchrotron crystallography.
Nat. Methods, 15, 2018
|
|
5ZHP
 
 | | M3 muscarinic acetylcholine receptor in complex with a selective antagonist | | Descriptor: | (1R,2R,4S,5S,7s)-7-({[4-fluoro-2-(thiophen-2-yl)phenyl]carbamoyl}oxy)-9,9-dimethyl-3-oxa-9-azatricyclo[3.3.1.0~2,4~]nonan-9-ium, CITRIC ACID, HEXAETHYLENE GLYCOL, ... | | Authors: | Liu, H, Hofmann, J, Fish, I, Schaake, B, Eitel, K, Bartuschat, A, Kaindl, J, Rampp, H, Banerjee, A, Hubner, H, Clark, M.J, Vincent, S.G, Fisher, J, Heinrich, M, Hirata, K, Liu, X, Sunahara, R.K, Shoichet, B.K, Kobilka, B.K, Gmeiner, P. | | Deposit date: | 2018-03-13 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-guided development of selective M3 muscarinic acetylcholine receptor antagonists
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5ZI7
 
 | |
3QI3
 
 | | Crystal structure of PDE9A(Q453E) in complex with inhibitor BAY73-6691 | | Descriptor: | 1-(2-chlorophenyl)-6-[(2R)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Hou, J, Xu, J, Liu, M, Zhao, R, Lou, H, Ke, H. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural asymmetry of phosphodiesterase-9, potential protonation of a glutamic Acid, and role of the invariant glutamine.
Plos One, 6, 2011
|
|
6FO0
 
 | | CryoEM structure of bovine cytochrome bc1 in complex with the anti-malarial compound GSK932121 | | Descriptor: | 3-chloro-6-(hydroxymethyl)-2-methyl-5-{4-[3-(trifluoromethoxy)phenoxy]phenyl}pyridin-4-ol, Chain I/V, Cytochrome b, ... | | Authors: | Johnson, R.M, Amporndanai, K, O'Neill, P.M, Fishwick, C.W.G, Jamson, A.H, Rawson, S.D, Hasnain, S.S, Antonyuk, S.V, Muench, S.P. | | Deposit date: | 2018-02-05 | | Release date: | 2018-02-28 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | X-ray and cryo-EM structures of inhibitor-bound cytochromebc1complexes for structure-based drug discovery.
IUCrJ, 5, 2018
|
|
5ZK7
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, ACE-ARG-TYR-SER-ARG-MK8-GLN-LEU-LEU-MK8-LEU-PHE-ARG-NH2, CHLORIDE ION, ... | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, F, Uhring, M. | | Deposit date: | 2018-03-23 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
3QE7
 
 | | Crystal Structure of Uracil Transporter--UraA | | Descriptor: | URACIL, Uracil permease, nonyl beta-D-glucopyranoside | | Authors: | Lu, F.R, Li, S, Yan, N. | | Deposit date: | 2011-01-20 | | Release date: | 2011-03-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.781 Å) | | Cite: | Structure and mechanism of the uracil transporter UraA
Nature, 472, 2011
|
|
5ZOA
 
 | |
4KI0
 
 | | Crystal structure of the maltose-binding protein/maltose transporter complex in an outward-facing conformation bound to maltohexaose | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, ABC transporter related protein, Binding-protein-dependent transport systems inner membrane component, ... | | Authors: | Oldham, M.L, Chen, S, Chen, J. | | Deposit date: | 2013-05-01 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for substrate specificity in the Escherichia coli maltose transport system.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
5ZML
 
 | | Stapled-peptides tailored against initiation of translation | | Descriptor: | 1,2-ETHANEDIOL, ACE-LYS-LYS-ARG-TYR-SER-ARG-MK8-GLN-LEU-LEU-MK8-PHE-ARG-ARG, Eukaryotic translation initiation factor 4E, ... | | Authors: | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Yano, J. | | Deposit date: | 2018-04-04 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
6GEO
 
 | |
4KJW
 
 | |
3O7U
 
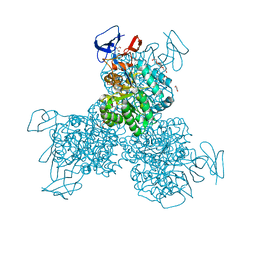 | | Crystal structure of Cytosine Deaminase from Escherichia Coli complexed with zinc and phosphono-cytosine | | Descriptor: | (2R)-2-amino-2,5-dihydro-1,5,2-diazaphosphinin-6(1H)-one 2-oxide, (2S)-1-[3-{[(2R)-2-hydroxypropyl]oxy}-2,2-bis({[(2R)-2-hydroxypropyl]oxy}methyl)propoxy]propan-2-ol, Cytosine deaminase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Hall, R.S, Raushel, F.M, Almo, S.C. | | Deposit date: | 2010-07-31 | | Release date: | 2011-06-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Three-dimensional structure and catalytic mechanism of Cytosine deaminase.
Biochemistry, 50, 2011
|
|
5WXF
 
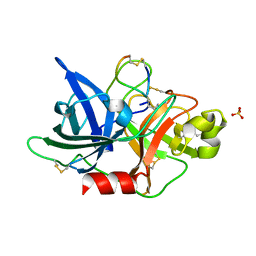 | | Crystal structure of uPA in complex with upain-2-2 | | Descriptor: | SULFATE ION, Urokinase-type plasminogen activator chain B, upain-2-2 peptide | | Authors: | Jiang, L, Huang, M. | | Deposit date: | 2017-01-07 | | Release date: | 2018-07-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Cleavage of peptidic inhibitors by target protease is caused by peptide conformational transition.
Biochim. Biophys. Acta, 1862, 2018
|
|
6GEQ
 
 | |
5WY8
 
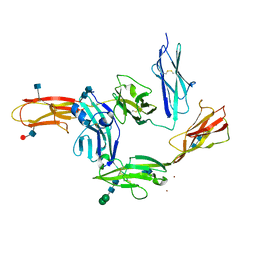 | | Crystal structure of PTP delta Ig1-Ig3 in complex with IL1RAPL1 Ig1-Ig3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-1 receptor accessory protein-like 1, ... | | Authors: | Kim, H.M, Won, S.Y, Kim, D. | | Deposit date: | 2017-01-11 | | Release date: | 2017-11-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | LAR-RPTP Clustering Is Modulated by Competitive Binding between Synaptic Adhesion Partners and Heparan Sulfate
Front Mol Neurosci, 10, 2017
|
|
4KMY
 
 | | Human folate receptor beta (FOLR2) at neutral pH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Folate receptor beta, POTASSIUM ION | | Authors: | Wibowo, A.S, Dann III, C.E. | | Deposit date: | 2013-05-08 | | Release date: | 2013-08-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Structures of human folate receptors reveal biological trafficking states and diversity in folate and antifolate recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4K5I
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with (R)-1,2-bis((2-amino-4-methylpyridin-6-yl)-methoxy)-propan-3-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-{[(2R)-3-aminopropane-1,2-diyl]bis(oxymethanediyl)}bis(4-methylpyridin-2-amine), ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-04-14 | | Release date: | 2013-09-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Chiral linkers to improve selectivity of double-headed neuronal nitric oxide synthase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3O9P
 
 | | The structure of the Escherichia coli murein tripeptide binding protein MppA | | Descriptor: | L-ALA-GAMMA-D-GLU-MESO-DIAMINOPIMELIC ACID, Periplasmic murein peptide-binding protein, ZINC ION | | Authors: | Maqbool, A, Levdikov, V.M, Blagova, E.V, Wilkinson, A.J, Thomas, G.H. | | Deposit date: | 2010-08-04 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Compensating Stereochemical Changes Allow Murein Tripeptide to Be Accommodated in a Conventional Peptide-binding Protein.
J.Biol.Chem., 286, 2011
|
|
