4WWX
 
 | | Crystal structure of the core RAG1/2 recombinase | | Descriptor: | V(D)J recombination-activating protein 1, V(D)J recombination-activating protein 2, ZINC ION | | Authors: | Kim, M.S, Lapkouski, M, Yang, W, Gellert, M. | | Deposit date: | 2014-11-12 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2001 Å) | | Cite: | Crystal structure of the V(D)J recombinase RAG1-RAG2.
Nature, 518, 2015
|
|
8R1G
 
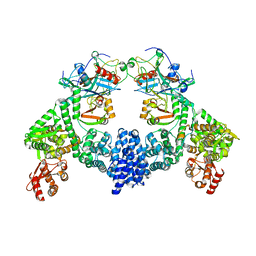 | | Dimeric ternary structure of E6AP-E6-p53 | | Descriptor: | Cellular tumor antigen p53, Ubiquitin-like protein SMT3,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Sandate, C.R, Chakrabory, D, Kater, L, Kempf, G, Thoma, N.H. | | Deposit date: | 2023-11-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structural insights into viral hijacking of p53 by E6 and E6AP
Biorxiv, 2023
|
|
8R1F
 
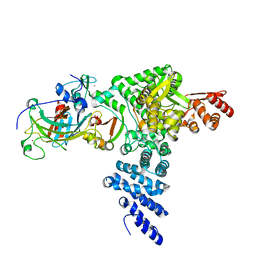 | | Monomeric E6AP-E6-p53 ternary complex | | Descriptor: | Cellular tumor antigen p53, Ubiquitin-like protein SMT3,Protein E6, Ubiquitin-protein ligase E3A, ... | | Authors: | Sandate, C.R, Chakraborty, D, Kater, L, Kempf, G, Thoma, N.H. | | Deposit date: | 2023-11-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Structural insights into viral hijacking of p53 by E6 and E6AP
Biorxiv, 2023
|
|
4MDN
 
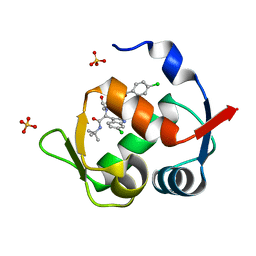 | | Structure of a novel submicromolar MDM2 inhibitor | | Descriptor: | 3-{(1S)-2-(tert-butylamino)-1-[{4-[(4-chlorobenzyl)oxy]benzyl}(formyl)amino]-2-oxoethyl}-6-chloro-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Bista, M, Popowicz, G, Holak, T.A. | | Deposit date: | 2013-08-23 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.905 Å) | | Cite: | Transient Protein States in Designing Inhibitors of the MDM2-p53 Interaction.
Structure, 21, 2013
|
|
7ML0
 
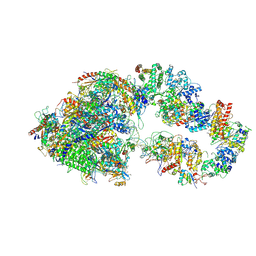 | | RNA polymerase II pre-initiation complex (PIC1) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML1
 
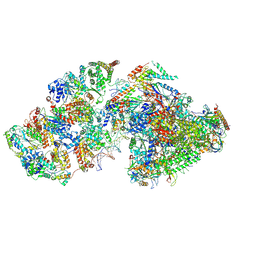 | | RNA polymerase II pre-initiation complex (PIC2) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML2
 
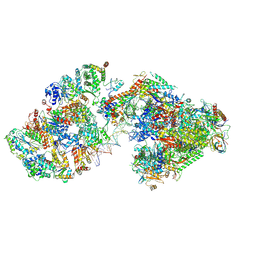 | | RNA polymerase II pre-initiation complex (PIC3) | | Descriptor: | BJ4_G0004860.mRNA.1.CDS.1, BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
7ML4
 
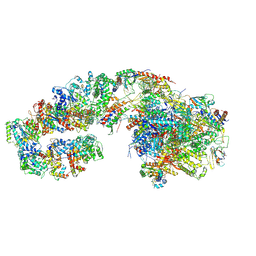 | | RNA polymerase II initially transcribing complex (ITC) | | Descriptor: | BJ4_G0050160.mRNA.1.CDS.1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB3, ... | | Authors: | Yang, C, Fujiwara, R, Kim, H.J, Gorbea Colon, J.J, Steimle, S, Garcia, B.A, Murakami, K. | | Deposit date: | 2021-04-27 | | Release date: | 2022-02-02 | | Last modified: | 2022-03-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Mol.Cell, 82, 2022
|
|
5BMU
 
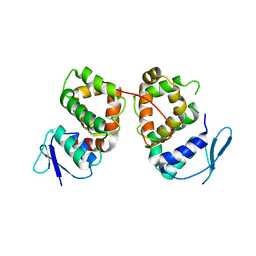 | |
7ABI
 
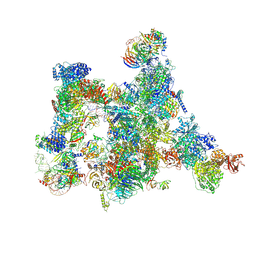 | | Human pre-Bact-2 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Beta-catenin-like protein 1, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
2PE6
 
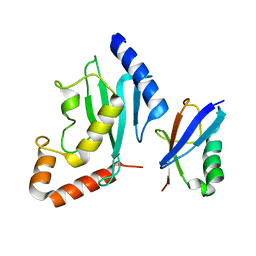 | | Non-covalent complex between human SUMO-1 and human Ubc9 | | Descriptor: | SUMO-conjugating enzyme UBC9, Small ubiquitin-related modifier 1 | | Authors: | Capili, A.D, Lima, C.D. | | Deposit date: | 2007-04-02 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Analysis of a Complex between SUMO and Ubc9 Illustrates Features of a Conserved E2-Ubl Interaction.
J.Mol.Biol., 369, 2007
|
|
6Q6G
 
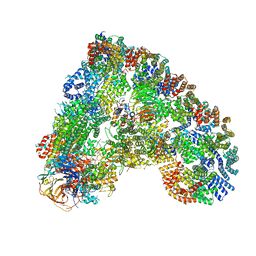 | | Cryo-EM structure of the APC/C-Cdc20-Cdk2-cyclinA2-Cks2 complex, the D1 box class | | Descriptor: | Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1, Anaphase-promoting complex subunit 10, Anaphase-promoting complex subunit 11, ... | | Authors: | Zhang, S, Barford, D. | | Deposit date: | 2018-12-11 | | Release date: | 2019-09-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cyclin A2 degradation during the spindle assembly checkpoint requires multiple binding modes to the APC/C.
Nat Commun, 10, 2019
|
|
6Q6H
 
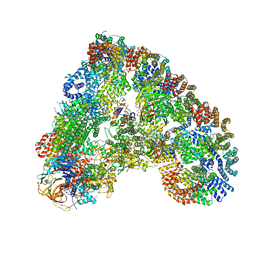 | |
8I0P
 
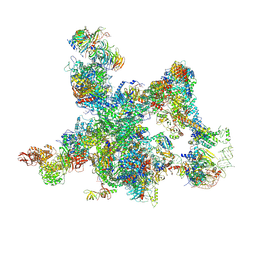 | | The cryo-EM structure of human pre-Bact complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
4R3Z
 
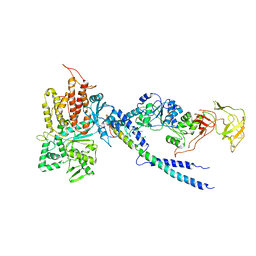 | | Crystal structure of human ArgRS-GlnRS-AIMP1 complex | | Descriptor: | Aminoacyl tRNA synthase complex-interacting multifunctional protein 1, Arginine--tRNA ligase, cytoplasmic, ... | | Authors: | Fu, Y, Kim, Y, Cho, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (4.033 Å) | | Cite: | Structure of the ArgRS-GlnRS-AIMP1 complex and its implications for mammalian translation
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7QVP
 
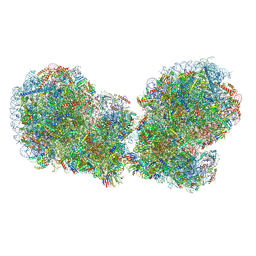 | | Human collided disome (di-ribosome) stalled on XBP1 mRNA | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Denk, T.G, Tesina, P, Beckmann, R. | | Deposit date: | 2022-01-22 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A distinct mammalian disome collision interface harbors K63-linked polyubiquitination of uS10 to trigger hRQT-mediated subunit dissociation.
Nat Commun, 13, 2022
|
|
8I0T
 
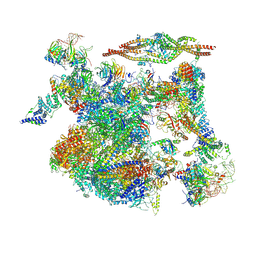 | | The cryo-EM structure of human Bact-III complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0R
 
 | | The cryo-EM structure of human Bact-I complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, BUD13 homolog, Cell division cycle 5-like protein, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
8I0S
 
 | | The cryo-EM structure of human Bact-II complex | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, Crooked neck-like protein 1, ... | | Authors: | Zhan, X, Lu, Y, Shi, Y. | | Deposit date: | 2023-01-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Molecular basis for the activation of human spliceosome.
Nat Commun, 15, 2024
|
|
4UI9
 
 | | Atomic structure of the human Anaphase-Promoting Complex | | Descriptor: | ANAPHASE-PROMOTING COMPLEX SUBUNIT 1, ANAPHASE-PROMOTING COMPLEX SUBUNIT 10, ANAPHASE-PROMOTING COMPLEX SUBUNIT 11, ... | | Authors: | Chang, L, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-17 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic Structure of the Apc and its Mechanism of Protein Ubiquitination
Nature, 522, 2015
|
|
7QTT
 
 | |
8CX2
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 2 | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
8CX0
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC monomeric complex | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
8CX1
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 1 | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
9B5G
 
 | |
