9GGL
 
 | | Cryo-EM structure of KBTBD4 WT-HDAC2 2:1 complex mediated by molecular glue UM171 | | Descriptor: | (1r,4r)-N~1~-[(7P)-2-benzyl-7-(2-methyl-2H-tetrazol-5-yl)-9H-pyrimido[4,5-b]indol-4-yl]cyclohexane-1,4-diamine, Histone deacetylase 2, Isoform 1 of Kelch repeat and BTB domain-containing protein 4, ... | | Authors: | Chen, Z, Chi, G, Pike, A.C.W, Montes, B, Bullock, A.N. | | Deposit date: | 2024-08-13 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural mimicry of UM171 and neomorphic cancer mutants co-opts E3 ligase KBTBD4 for HDAC1/2 recruitment.
Nat Commun, 16, 2025
|
|
5IQ9
 
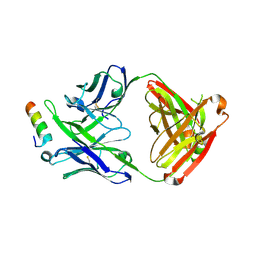 | | Crystal structure of 10E8v4 Fab in complex with an HIV-1 gp41 peptide. | | Descriptor: | 10E8v4 Heavy Chain, 10E8v4 Light Chain, gp41 MPER peptide | | Authors: | Ofek, G, Kwon, Y.D, Caruso, W, Kwong, P.D. | | Deposit date: | 2016-03-10 | | Release date: | 2016-04-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Optimization of the Solubility of HIV-1-Neutralizing Antibody 10E8 through Somatic Variation and Structure-Based Design.
J.Virol., 90, 2016
|
|
5CTA
 
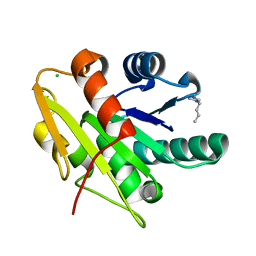 | | G158E/K44E/R57E/Y49E Bacillus subtilis lipase A with 10% [BMIM][Cl] | | Descriptor: | 1-butyl-3-methyl-1H-imidazol-3-ium, CHLORIDE ION, Esterase | | Authors: | Nordwald, E.M, Plaks, J.G, Snell, J.R, Sousa, M.C, Kaar, J.L. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystallographic Investigation of Imidazolium Ionic Liquid Effects on Enzyme Structure.
Chembiochem, 16, 2015
|
|
5H6J
 
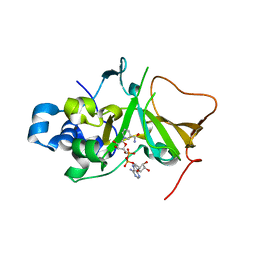 | | DNA targeting ADP-ribosyltransferase Pierisin-1 in complex with beta-NAD+ | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Pierisin-1 | | Authors: | Oda, T, Hirabayashi, H, Shikauchi, G, Takamura, R, Hiraga, K, Minami, H, Hashimoto, H, Yamamoto, M, Wakabayashi, K, Sugimura, T, Shimizu, T, Sato, M. | | Deposit date: | 2016-11-14 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of autoinhibition and activation of the DNA-targeting ADP-ribosyltransferase pierisin-1
J. Biol. Chem., 292, 2017
|
|
7ZVA
 
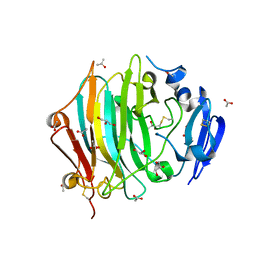 | | Crystal Structure of the native zymogen form of the glutamic-class prolyl-endopeptidase neprosin at 1.80 A resolution. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, C-terminal peptidase, ... | | Authors: | Del Amo-Maestro, L, Eckhard, U, Rodriguez-Banqueri, A, Mendes, S.R, Guevara, T, Gomis-Ruth, F.X. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular and in vivo studies of a glutamate-class prolyl-endopeptidase for coeliac disease therapy.
Nat Commun, 13, 2022
|
|
6LFD
 
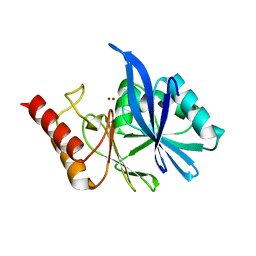 | |
7ZQS
 
 | | Cryo-EM Structure of Human Transferrin Receptor 1 bound to DNA Aptamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (30-MER), ... | | Authors: | Bansia, H, Wang, T, Gutierrez, D, des Georges, A. | | Deposit date: | 2022-05-02 | | Release date: | 2022-08-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Discovery of a Transferrin Receptor 1-Binding Aptamer and Its Application in Cancer Cell Depletion for Adoptive T-Cell Therapy Manufacturing.
J.Am.Chem.Soc., 144, 2022
|
|
6L14
 
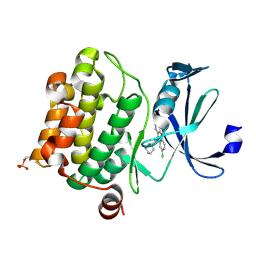 | | Crystal structure of Ser/Thr kinase Pim1 in complex with 10-DEBC derivatives | | Descriptor: | 2-chloranyl-10-[3-[(3~{S})-piperidin-3-yl]propyl]phenoxazine, Serine/threonine-protein kinase pim-1 | | Authors: | Zhang, W, Xie, Y, Cao, R, Huang, N, Zhou, Y. | | Deposit date: | 2019-09-27 | | Release date: | 2020-05-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Optimization of 10-DEBC Derivatives as Potent and Selective Pim-1 Kinase Inhibitors.
J.Chem.Inf.Model., 60, 2020
|
|
6C2Z
 
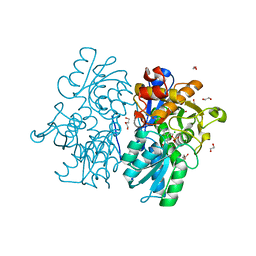 | | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: the Structure of the PLP-Aminoacrylate Intermediate | | Descriptor: | 1,2-ETHANEDIOL, 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CALCIUM ION, ... | | Authors: | Kreinbring, C.A, Tu, Y, Liu, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2018-01-09 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal Structures of Cystathionine beta-Synthase from Saccharomyces cerevisiae: One Enzymatic Step at a Time.
Biochemistry, 57, 2018
|
|
5DYF
 
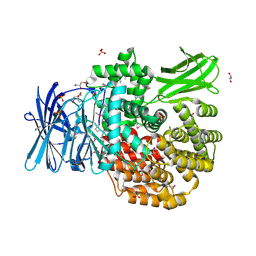 | | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid | | Descriptor: | Aminopeptidase N, GLYCEROL, IMIDAZOLE, ... | | Authors: | Nocek, B, Joachimiak, A, Vassiliou, S, Berlicki, L, Mucha, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-09-24 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | The crystal structure of Aminopeptidase N in complex with N-benzyl-1,2-diaminoethylphosphonic acid
To Be Published
|
|
7ZSA
 
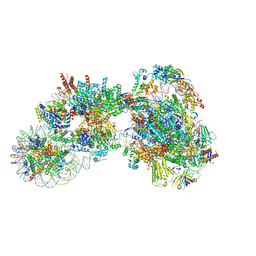 | |
4AZG
 
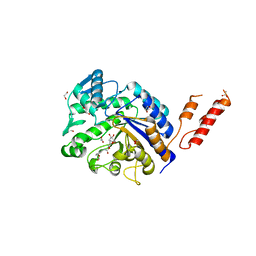 | | Differential inhibition of the tandem GH20 catalytic modules in the pneumococcal exo-beta-D-N-acetylglucosaminidase, StrH | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BETA-N-ACETYLHEXOSAMINIDASE, ... | | Authors: | Pluvinage, B, Stubbs, K.A, Vocadlo, D.J, Boraston, A.B. | | Deposit date: | 2012-06-25 | | Release date: | 2013-07-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Inhibition of the Family 20 Glycoside Hydrolase Catalytic Modules in the Streptococcus Pneumoniae Exo-Beta-D-N-Acetylglucosaminidase, Strh.
Org.Biomol.Chem., 11, 2013
|
|
5U1P
 
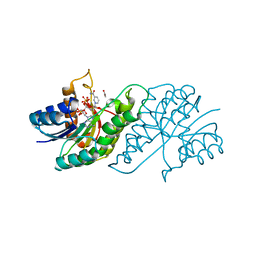 | |
6ZIS
 
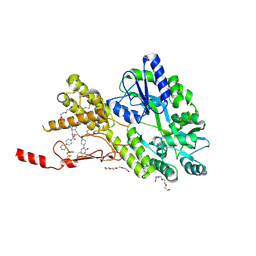 | | Crystal structure of a CGRP receptor ectodomain heterodimer with bound high affinity inhibitor | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Receptor activity-modifying protein 1,Calcitonin gene-related peptide type 1 receptor, N-{(1S)-5-amino-1-[(4-pyridin-4-ylpiperazin-1-yl)carbonyl]pentyl}-3,5-dibromo-Nalpha-{[4-(2-oxo-1,4-dihydroquinazolin-3 (2H)-yl)piperidin-1-yl]carbonyl}-D-tyrosinamide, TETRAETHYLENE GLYCOL, ... | | Authors: | Southall, S.M. | | Deposit date: | 2020-06-26 | | Release date: | 2020-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Based Drug Discovery ofN-((R)-3-(7-Methyl-1H-indazol-5-yl)-1-oxo-1-(((S)-1-oxo-3-(piperidin-4-yl)-1-(4-(pyridin-4-yl)piperazin-1-yl)propan-2-yl)amino)propan-2-yl)-2'-oxo-1',2'-dihydrospiro[piperidine-4,4'-pyrido[2,3-d][1,3]oxazine]-1-carboxamide (HTL22562): A Calcitonin Gene-Related Peptide Receptor Antagonist for Acute Treatment of Migraine.
J.Med.Chem., 63, 2020
|
|
7ZS9
 
 | |
8A1Z
 
 | | Crystal structure of Phosphoserine phosphatase SerB from Mycobacterium avium in complex with 1-(2,4-dichlorophenyl)-3-hydroxyurea | | Descriptor: | 1-(2,4-dichlorophenyl)-3-oxidanyl-urea, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Haufroid, M, Wouters, J. | | Deposit date: | 2022-06-02 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Targeting the phosphoserine phosphatase MtSerB2 for tuberculosis drug discovery, an hybrid knowledge based /fragment based approach.
Eur.J.Med.Chem., 245, 2022
|
|
9CF8
 
 | |
5A6J
 
 | | GH20C, Beta-hexosaminidase from Streptococcus pneumoniae | | Descriptor: | 1,2-ETHANEDIOL, N-ACETYL-BETA-D-GLUCOSAMINIDASE | | Authors: | Cid, M, Robb, C.S, Higgins, M.A, Boraston, A.B. | | Deposit date: | 2015-06-26 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Second beta-Hexosaminidase Encoded in the Streptococcus pneumoniae Genome Provides an Expanded Biochemical Ability to Degrade Host Glycans.
J. Biol. Chem., 290, 2015
|
|
7T7T
 
 | |
6GB1
 
 | | Crystal structure of the GLP1 receptor ECD with Peptide 11 | | Descriptor: | Glucagon-like peptide 1 receptor, HEXANE-1,6-DIOL, Peptide 11, ... | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2018-04-13 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Dual Glucagon-like Peptide 1 (GLP-1)/Glucagon Receptor Agonists Specifically Optimized for Multidose Formulations.
J. Med. Chem., 61, 2018
|
|
6S55
 
 | |
7MKC
 
 | |
2V0W
 
 | |
8WE6
 
 | | Human L-type voltage-gated calcium channel Cav1.2 at 2.9 Angstrom resolution | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
6RX0
 
 | | Trypanosoma brucei PTR1 (TbPTR1) in complex with inhibitor 3 (NMT-C0013) | | Descriptor: | ACETATE ION, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Landi, G, Pozzi, C, Mangani, S. | | Deposit date: | 2019-06-07 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Multitarget, Selective Compound Design Yields Potent Inhibitors of a Kinetoplastid Pteridine Reductase 1.
J.Med.Chem., 65, 2022
|
|
