3U4N
 
 | | A novel covalently linked insulin dimer | | Descriptor: | Insulin A chain, Insulin B chain | | Authors: | Norrman, M, Vinther, T.N. | | Deposit date: | 2011-10-10 | | Release date: | 2012-04-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Novel covalently linked insulin dimer engineered to investigate the function of insulin dimerization.
Plos One, 7, 2012
|
|
3QK6
 
 | | Crystal structure of Escherichia coli PhnD | | Descriptor: | PhnD, subunit of alkylphosphonate ABC transporter, UNKNOWN LIGAND | | Authors: | Alicea, I, Schreiter, E.R. | | Deposit date: | 2011-01-31 | | Release date: | 2011-10-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Escherichia coli Phosphonate Binding Protein PhnD and Rationally Optimized Phosphonate Biosensors.
J.Mol.Biol., 414, 2011
|
|
1IP9
 
 | | SOLUTION STRUCTURE OF THE PB1 DOMAIN OF BEM1P | | Descriptor: | BEM1 PROTEIN | | Authors: | Terasawa, H, Noda, Y, Ito, T, Hatanaka, H, Ichikawa, S, Ogura, K, Sumimoto, H, Inagaki, F. | | Deposit date: | 2001-04-26 | | Release date: | 2001-08-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and ligand recognition of the PB1 domain: a novel protein module binding to the PC motif.
EMBO J., 20, 2001
|
|
3OWZ
 
 | | Crystal structure of glycine riboswitch, soaked in Iridium | | Descriptor: | Domain II of glycine riboswitch, GLYCINE, IRIDIUM HEXAMMINE ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-20 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.949 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
3D4J
 
 | | Crystal structure of Human mevalonate diphosphate decarboxylase | | Descriptor: | Diphosphomevalonate decarboxylase, SULFATE ION | | Authors: | Voynova, N.E, Fu, Z, Battaile, K, Herdendorf, T.J, Kim, J.-J.P, Miziorko, H.M. | | Deposit date: | 2008-05-14 | | Release date: | 2008-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human mevalonate diphosphate decarboxylase: characterization, investigation of the mevalonate diphosphate binding site, and crystal structure.
Arch.Biochem.Biophys., 480, 2008
|
|
3OXB
 
 | |
3OXM
 
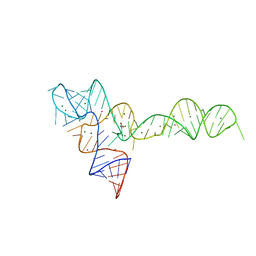 | | crystal structure of glycine riboswitch, Tl-Acetate soaked | | Descriptor: | GLYCINE, MAGNESIUM ION, THALLIUM (I) ION, ... | | Authors: | Huang, L, Serganov, A, Patel, D.J. | | Deposit date: | 2010-09-21 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch.
Mol.Cell, 40, 2010
|
|
2D1N
 
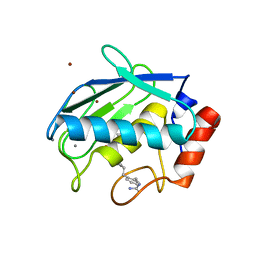 | | Collagenase-3 (MMP-13) complexed to a hydroxamic acid inhibitor | | Descriptor: | CALCIUM ION, Collagenase 3, SM-25453, ... | | Authors: | Kohno, T, Hochigai, H, Yamashita, E, Tsukihara, T, Kanaoka, M. | | Deposit date: | 2005-08-29 | | Release date: | 2006-06-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structures of the catalytic domain of human stromelysin-1 (MMP-3) and collagenase-3 (MMP-13) with a hydroxamic acid inhibitor SM-25453
Biochem.Biophys.Res.Commun., 344, 2006
|
|
1TSD
 
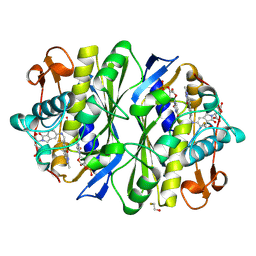 | |
1TT8
 
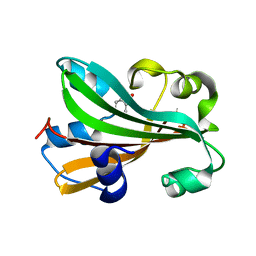 | | CHORISMATE LYASE WITH PRODUCT, 1.0 A RESOLUTION | | Descriptor: | Chorismate-pyruvate lyase, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M.J, Vilker, V, Howard, A. | | Deposit date: | 2004-06-22 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural analysis of ligand binding and catalysis in chorismate lyase.
Arch.Biochem.Biophys., 445, 2006
|
|
2Z7Q
 
 | |
6X2O
 
 | | Crystal Structure of unliganded CRM1(E571K)-Ran-RanBP1 | | Descriptor: | Exportin-1, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Baumhardt, J.M. | | Deposit date: | 2020-05-20 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.551 Å) | | Cite: | Recognition of nuclear export signals by CRM1 carrying the oncogenic E571K mutation.
Mol.Biol.Cell, 31, 2020
|
|
1UJR
 
 | | Solution structure of WWE domain in BAB28015 | | Descriptor: | hypothetical protein AK012080 | | Authors: | He, F, Muto, Y, Hamana, H, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nagayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-08-11 | | Release date: | 2004-10-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of WWE domain in BAB28015
To be Published
|
|
3L3P
 
 | | Crystal structure of the C-terminal domain of Shigella type III effector IpaH9.8, with a novel domain swap | | Descriptor: | IpaH9.8 | | Authors: | Seyedarabi, A, Sullivan, J.A, Sasakawa, C, Pickersgill, R.W. | | Deposit date: | 2009-12-17 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A disulfide driven domain swap switches off the activity of Shigella IpaH9.8 E3 ligase
Febs Lett., 584, 2010
|
|
8F47
 
 | |
1UKX
 
 | | Solution structure of the RWD domain of mouse GCN2 | | Descriptor: | GCN2 eIF2alpha kinase | | Authors: | Nameki, N, Yoneyama, M, Koshiba, S, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-03 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the RWD domain of the mouse GCN2 protein.
Protein Sci., 13, 2004
|
|
5E6X
 
 | |
3U2Q
 
 | | EF-Tu (Escherichia coli) in complex with NVP-LFF571 | | Descriptor: | Elongation factor Tu 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Palestrant, D.J. | | Deposit date: | 2011-10-04 | | Release date: | 2012-05-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of LFF571: an investigational agent for Clostridium difficile infection.
J.Med.Chem., 55, 2012
|
|
6U75
 
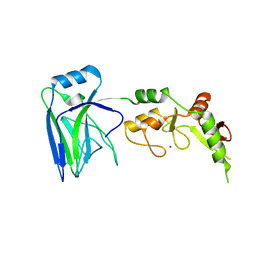 | |
6UD7
 
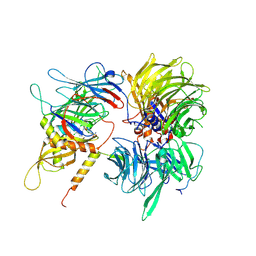 | | Crystal structure of full-length human DCAF15-DDB1(deltaBPB)-DDA1-RBM39 in complex with indisulam | | Descriptor: | DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, DNA damage-binding protein 1,DNA damage-binding protein 1, ... | | Authors: | Bussiere, D.E, Shu, W, Xie, L, Knapp, M. | | Deposit date: | 2019-09-18 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.
Nat.Chem.Biol., 16, 2020
|
|
8USP
 
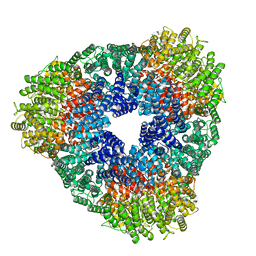 | |
8USQ
 
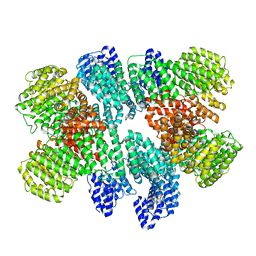 | |
5E6V
 
 | |
4QIR
 
 | | Crystal structure of Aminopeptidase N in complex with the phosphinic dipeptide analogue LL-(R,S)-2-(pyridin-3-yl)ethylGlyP[CH2]Phe | | Descriptor: | 3-{[(R)-1-amino-3-(pyridin-3-yl)propyl](hydroxy)phosphoryl}-(S)-2-benzylpropanoic acid, Aminopeptidase N, GLYCEROL, ... | | Authors: | Nocek, B, Joachimiak, A, Berlicki, L, Vassiliou, S, Mucha, A. | | Deposit date: | 2014-06-01 | | Release date: | 2014-09-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Structure-guided, single-point modifications in the phosphinic dipeptide structure yield highly potent and selective inhibitors of neutral aminopeptidases.
J.Med.Chem., 57, 2014
|
|
3A8G
 
 | | Crystal structure of Nitrile Hydratase mutant S113A complexed with Trimethylacetonitrile | | Descriptor: | 2,2-dimethylpropanenitrile, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Yamanaka, Y, Hashimoto, K, Ohtaki, A, Noguchi, K, Yohda, M, Odaka, M. | | Deposit date: | 2009-10-06 | | Release date: | 2010-04-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Kinetic and structural studies on roles of the serine ligand and a strictly conserved tyrosine residue in nitrile hydratase
J.Biol.Inorg.Chem., 15, 2010
|
|
