1QQE
 
 | |
5XGQ
 
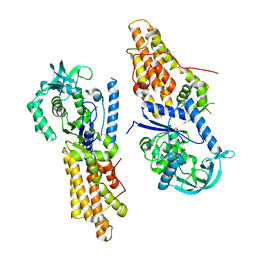 | |
6IRD
 
 | | Complex structure of INADL PDZ89 and PLCb4 C-terminal CC-PBM | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase, GOLD ION, InaD-like protein | | Authors: | Ye, F, Li, J, Huang, Y, Liu, W, Zhang, M. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.813 Å) | | Cite: | An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Elife, 7, 2018
|
|
5XH9
 
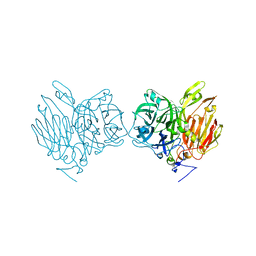 | | Aspergillus kawachii beta-fructofuranosidase | | Descriptor: | Extracellular invertase, SODIUM ION | | Authors: | Nagaya, M, Tonozuka, T. | | Deposit date: | 2017-04-19 | | Release date: | 2017-07-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a beta-fructofuranosidase with high transfructosylation activity from Aspergillus kawachii
Biosci. Biotechnol. Biochem., 81, 2017
|
|
5XJD
 
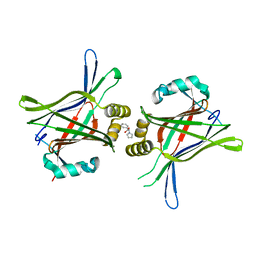 | | TEAD in complex with fragment | | Descriptor: | (2S)-2-phenyl-2-pyrrol-1-yl-ethanoic acid, Transcriptional enhancer factor TEF-3 | | Authors: | Kaan, H.Y.K, Sim, A.Y.L, Tan, S.K.J, Verma, C, Song, H. | | Deposit date: | 2017-05-01 | | Release date: | 2018-01-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Targeting YAP/TAZ-TEAD protein-protein interactions using fragment-based and computational modeling approaches.
PLoS ONE, 12, 2017
|
|
5XHM
 
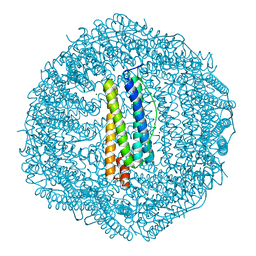 | | Crystal structure of Frog M-ferritin D40A mutant | | Descriptor: | CHLORIDE ION, Ferritin, middle subunit, ... | | Authors: | Jagdev, M.K, Vasudevan, D. | | Deposit date: | 2017-04-21 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Surface charge dependent separation of modified and hybrid ferritin in native PAGE: Impact of lysine 104
Biochim. Biophys. Acta, 1865, 2017
|
|
1QSN
 
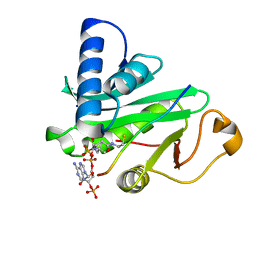 | | CRYSTAL STRUCTURE OF TETRAHYMENA GCN5 WITH BOUND COENZYME A AND HISTONE H3 PEPTIDE | | Descriptor: | COENZYME A, HISTONE H3, TGCN5 HISTONE ACETYL TRANSFERASE | | Authors: | Rojas, J.R, Trievel, R.C, Zhou, J, Mo, Y, Li, X, Berger, S.L, David Allis, C, Marmorstein, R. | | Deposit date: | 1999-06-22 | | Release date: | 1999-09-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide.
Nature, 401, 1999
|
|
5XHS
 
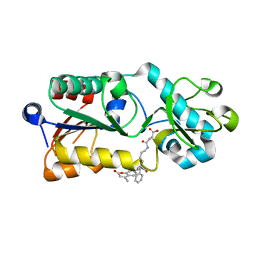 | | Crystal structure of SIRT5 complexed with a fluorogenic small-molecule substrate SuBKA | | Descriptor: | (2S)-2-azanyl-6-[(4-hydroxy-4-oxo-butanoyl)amino]hexanoic acid, 7-AMINO-4-METHYL-CHROMEN-2-ONE, NAD-dependent protein deacylase sirtuin-5, ... | | Authors: | Yu, Y, Li, B, Chen, Q. | | Deposit date: | 2017-04-24 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Interactions between sirtuins and fluorogenic small-molecule substrates offer insights into inhibitor design
Rsc Adv, 7, 2017
|
|
1QSQ
 
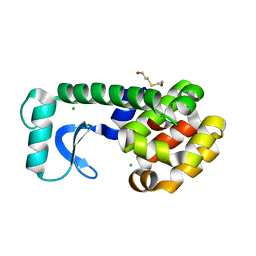 | | CAVITY CREATING MUTATION | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J, Matthews, B.W. | | Deposit date: | 1999-06-22 | | Release date: | 1999-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
6I2B
 
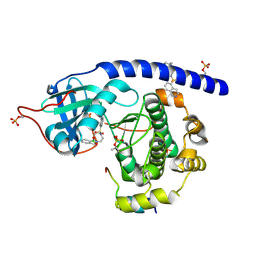 | | Crystal Structure of the Protein-Kinase A catalytic subunit from Cricetulus Griseus in complex with compounds RKp153 and RKp117 | | Descriptor: | UPF0418 protein FAM164A, [2-[(4-isoquinolin-5-ylsulfonyl-1,4-diazepan-1-yl)methyl]phenyl]boronic acid, beta-D-ribopyranose, ... | | Authors: | Mueller, J.M, Heine, A, Klebe, G. | | Deposit date: | 2018-11-01 | | Release date: | 2019-05-15 | | Last modified: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Conceptional Design of Self-Assembling Bisubstrate-like Inhibitors of Protein Kinase A Resulting in a Boronic Acid Glutamate Linkage
Acs Omega, 2019
|
|
5XK3
 
 | | Crystal structure of apo form Isosesquilavandulyl Diphosphate Synthase from Streptomyces sp. strain CNH-189 | | Descriptor: | SULFATE ION, Undecaprenyl diphosphate synthase | | Authors: | Ko, T.P, Guo, R.T, Liu, W, Chen, C.C, Gao, J. | | Deposit date: | 2017-05-05 | | Release date: | 2018-01-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | "Head-to-Middle" and "Head-to-Tail" cis-Prenyl Transferases: Structure of Isosesquilavandulyl Diphosphate Synthase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
1QSJ
 
 | | N-TERMINALLY TRUNCATED C3DG FRAGMENT | | Descriptor: | COMPLEMENT C3 PRECURSOR | | Authors: | Zanotti, G, Bassetto, A, Battistutta, R, Stoppini, M, Folli, C, Berni, R. | | Deposit date: | 1999-06-22 | | Release date: | 2000-07-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure at 1.44 A resolution of an N-terminally truncated form of the rat serum complement C3d fragment.
Biochim.Biophys.Acta, 1478, 2000
|
|
1QTB
 
 | |
6I2K
 
 | | Structure of EV71 complexed with its receptor SCARB2 | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhou, D, Zhao, Y, Kotecha, A, Fry, E.E, Kelly, J, Wang, X, Rao, Z, Rowlands, D.J, Ren, J, Stuart, D.I. | | Deposit date: | 2018-11-01 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Unexpected mode of engagement between enterovirus 71 and its receptor SCARB2.
Nat Microbiol, 4, 2019
|
|
1QTP
 
 | |
6I8Y
 
 | | Crystal structure of Spindlin1 in complex with the Methyltransferase inhibitor A366 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 5'-methoxy-6'-[3-(pyrrolidin-1-yl)propoxy]spiro[cyclobutane-1,3'-indol]-2'-amine, ... | | Authors: | Srikannathasan, V, Johansson, C, Gileadi, C, Shrestha, L, Sorrell, F.J, Krojer, T, Burgess-Brown, N.A, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U.C.T. | | Deposit date: | 2018-11-21 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | A Chemical Probe for Tudor Domain Protein Spindlin1 to Investigate Chromatin Function.
J.Med.Chem., 62, 2019
|
|
6I30
 
 | | Crystal structure of the AmpC from Pseudomonas aeruginosa with 1C | | Descriptor: | (3R)-3-(cyclohexylcarbonylamino)-2-oxidanyl-3,4-dihydro-1,2-benzoxaborinine-8-carboxylic acid, Class C beta-lactamase PDC-301 | | Authors: | Brem, J, Cahill, S.T, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2018-11-02 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Studies on the inhibition of AmpC and other beta-lactamases by cyclic boronates.
Biochim Biophys Acta Gen Subj, 1863, 2019
|
|
1QSW
 
 | |
5XJV
 
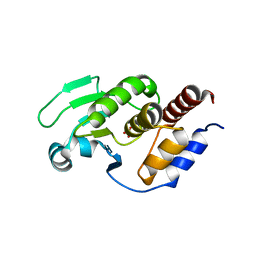 | | Two intermediate states of conformation switch in dual specificity phosphatase 13a | | Descriptor: | Dual specificity protein phosphatase 13 isoform A, PHOSPHATE ION | | Authors: | Wei, C.H, Min, H.G, Chun, H.J, Ryu, S.E. | | Deposit date: | 2017-05-04 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Two intermediate states of the conformational switch in dual specificity phosphatase 13a
Pharmacol. Res., 128, 2018
|
|
6I96
 
 | |
5XLK
 
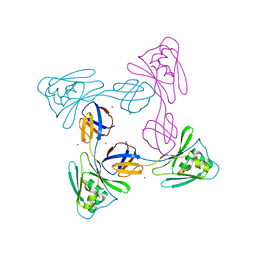 | | Crystal structure of the flagellar cap protein FliD D2-D3 domains from Serratia marcescens in Space group I422 | | Descriptor: | Flagellar hook-associated protein 2, ZINC ION | | Authors: | Cho, S.Y, Song, W.S, Hong, H.J, Yoon, S.I. | | Deposit date: | 2017-05-10 | | Release date: | 2017-06-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Tetrameric structure of the flagellar cap protein FliD from Serratia marcescens.
Biochem. Biophys. Res. Commun., 489, 2017
|
|
1QVA
 
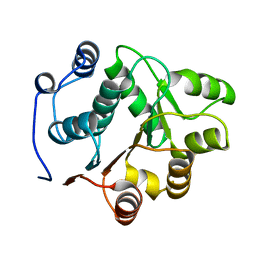 | | YEAST INITIATION FACTOR 4A N-TERMINAL DOMAIN | | Descriptor: | INITIATION FACTOR 4A | | Authors: | Johnson, E.R, McKay, D.B. | | Deposit date: | 1999-07-07 | | Release date: | 1999-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic structure of the amino terminal domain of yeast initiation factor 4A, a representative DEAD-box RNA helicase
RNA, 5, 1999
|
|
5XK2
 
 | |
1QU5
 
 | |
5XKE
 
 | | Crystal structure of T2R-TTL-Demecolcine complex | | Descriptor: | (7S)-1,2,3,10-tetramethoxy-7-(methylamino)-6,7-dihydro-5H-benzo[a]heptalen-9-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-07 | | Release date: | 2018-07-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Identification of a powerful and reversible microtubule-inhibitor with efficacy against multidrug-resistant tumors
To Be Published
|
|
