6W6T
 
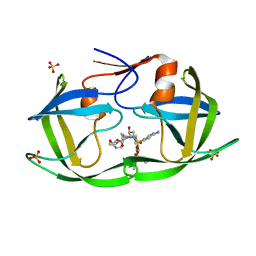 | | WT HIV-1 Protease in Complex with Phosphonated UMass6 (PU6) | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Nalivaika, E.A, Ali, A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2020-03-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | To Be Determined
To Be Published
|
|
6W6R
 
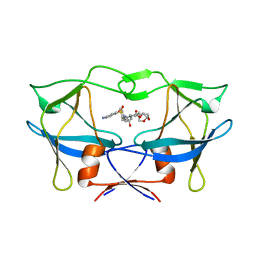 | | WT HTLV-1 Protease in Complex with UMass6 (UM6) | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl [(1S,2R)-3-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-1-benzyl-2-hydroxypropyl]carbamate, HTLV-1 Protease | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Nalivaika, E.A, Ali, A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2020-03-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | To Be Determined
To Be Published
|
|
6W6S
 
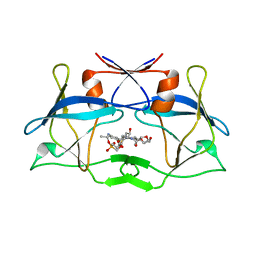 | | WT HTLV-1 Protease in Complex with Phosphonated UMass6 (PU6) | | Descriptor: | HTLV-1 Protease, diethyl [(4-{(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Henes, M, Kosovrasti, K, Nalivaika, E.A, Ali, A, KurtYilmaz, N, Schiffer, C.A. | | Deposit date: | 2020-03-17 | | Release date: | 2021-03-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | To Be Determined
To Be Published
|
|
6W6J
 
 | |
6W6I
 
 | |
6M76
 
 | | GH31 alpha-N-acetylgalactosaminidase from Enterococcus faecalis | | Descriptor: | 1,2-ETHANEDIOL, LPXTG-motif cell wall anchor domain protein | | Authors: | Miyazaki, T. | | Deposit date: | 2020-03-17 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of the Enterococcus faecalis alpha-N-acetylgalactosaminidase, a member of the glycoside hydrolase family 31.
Febs Lett., 594, 2020
|
|
6YB6
 
 | | Thrombin in complex with D-Phe-Pro-3-chloro-1,3-dihydroxybenzylamide derivative (13c) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D-Phe-Pro-3-chloro-1,3-dihydroxybenzylamide derivative, DIMETHYL SULFOXIDE, ... | | Authors: | Sandner, A, Heine, A, Klebe, G, Abazi, N. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-31 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Thrombin in complex with D-Phe-Pro-3-chloro-1,3-dihydroxybenzylamide derivative (13c)
To be published
|
|
6M6X
 
 | | Oridonin in complex with CRM1#-Ran-RanBP1 | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-03-16 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Novel Mechanistic Observations and NES-Binding Groove Features Revealed by the CRM1 Inhibitors Plumbagin and Oridonin.
J.Nat.Prod., 84, 2021
|
|
6W6G
 
 | |
6W6H
 
 | |
6W6E
 
 | |
6YAW
 
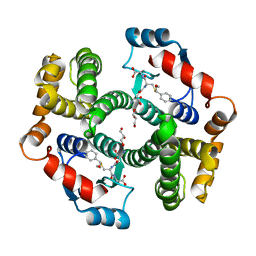 | | Crystal structure of human GSTA1-1 bound to the glutathione adduct of cinnamaldehyde | | Descriptor: | (2~{S})-2-azanyl-5-[[(2~{R})-1-(2-hydroxy-2-oxoethylamino)-1-oxidanylidene-3-[(1~{R})-3-oxidanylidene-1-phenyl-propyl]sulfanyl-propan-2-yl]amino]-5-oxidanylidene-pentanoic acid, GLYCEROL, Glutathione S-transferase A1 | | Authors: | Schwartz, M, Neiers, F. | | Deposit date: | 2020-03-13 | | Release date: | 2020-08-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Interactions Between Odorants and Glutathione Transferases in the Human Olfactory Cleft.
Chem.Senses, 45, 2020
|
|
6YAU
 
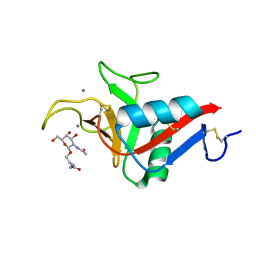 | | CRYSTAL STRUCTURE OF ASGPR 1 IN COMPLEX WITH GN-A. | | Descriptor: | 5-[(2~{R},3~{R},4~{R},5~{R},6~{R})-3-acetamido-6-(hydroxymethyl)-4,5-bis(oxidanyl)oxan-2-yl]oxy-~{N}-[3-(propanoylamino)propyl]pentanamide, Asialoglycoprotein receptor 1, CALCIUM ION | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2020-03-13 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.397 Å) | | Cite: | Triantennary GalNAc Molecular Imaging Probes for Monitoring Hepatocyte Function in a Rat Model of Nonalcoholic Steatohepatitis.
Adv Sci, 7, 2020
|
|
6YAT
 
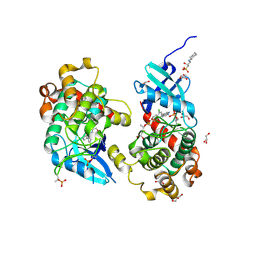 | | Crystal structure of STK4 (MST1) in complex with compound 6 | | Descriptor: | (2R)-3-(cyclohexylamino)-2-hydroxypropane-1-sulfonic acid, 4-[5-(3-chlorophenyl)-7~{H}-pyrrolo[2,3-d]pyrimidin-4-yl]morpholine, GLYCEROL, ... | | Authors: | Chaikuad, A, Bata, N, Limpert, A.S, Lambert, L.J, Bakas, N.A, Cosford, N.D.P, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-03-13 | | Release date: | 2020-04-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Inhibitors of the Hippo Pathway Kinases STK3/MST2 and STK4/MST1 Have Utility for the Treatment of Acute Myeloid Leukemia.
J.Med.Chem., 65, 2022
|
|
6YAL
 
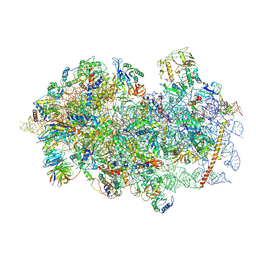 | | Mammalian 48S late-stage initiation complex with beta-globin mRNA | | Descriptor: | 18S ribosomal RNA, 40S Ribosomal protein uS3, 40S Ribosomal protein uS7, ... | | Authors: | Bochler, A, Simonetti, A, Guca, E, Hashem, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-04-08 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Insights into the Mammalian Late-Stage Initiation Complexes.
Cell Rep, 31, 2020
|
|
6M5X
 
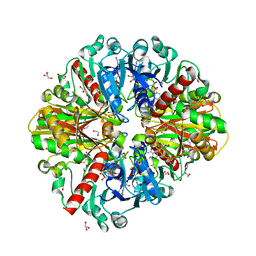 | | A fungal glyceraldehyde-3-phosphate dehydrogenase with self-resistance to inhibitor heptelidic acid | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yan, Y, Zang, X, Cooper, S.J, Lin, H, Zhou, J, Tang, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.05990934 Å) | | Cite: | Biosynthesis of the fungal glyceraldehyde-3-phosphate dehydrogenase inhibitor heptelidic acid and mechanism of self-resistance
Chem Sci, 11, 2020
|
|
6M63
 
 | | Crystal structure of a cAMP sensor G-Flamp1. | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Chimera of Cyclic nucleotide-gated potassium channel mll3241 and Yellow fluorescent protein | | Authors: | Zhou, Z, Chen, S, Wang, L, Chu, J. | | Deposit date: | 2020-03-12 | | Release date: | 2021-09-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A high-performance genetically encoded fluorescent indicator for in vivo cAMP imaging.
Nat Commun, 13, 2022
|
|
6M60
 
 | | Plumbagin in complex with CRM1#-Ran-RanBP1 | | Descriptor: | (2~{R})-2-methyl-5-oxidanyl-2,3-dihydronaphthalene-1,4-dione, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Lei, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Novel Mechanistic Observations and NES-Binding Groove Features Revealed by the CRM1 Inhibitors Plumbagin and Oridonin.
J.Nat.Prod., 84, 2021
|
|
6M62
 
 | |
6W50
 
 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR METHYL ((10R,14S)- 14-(4-(3-CHLORO-2,6-DIFLUOROPHENYL)-6-OXO-3,6-DIHYDRO- 1(2H)-PYRIDINYL)-10-METHYL-9-OXO-8,16- DIAZATRICYCLO[13.3.1.0~2,7~]NONADECA-1(19),2,4,6,15,17- HEXAEN-5-YL)CARBAMATE | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2020-03-12 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of a High Affinity, Orally Bioavailable Macrocyclic FXIa Inhibitor with Antithrombotic Activity in Preclinical Species.
J.Med.Chem., 63, 2020
|
|
6W5B
 
 | |
6M61
 
 | | Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) with inhibitor heptelidic acid | | Descriptor: | (5aS,6R,9S,9aS)-9-methyl-9-oxidanyl-1-oxidanylidene-6-propan-2-yl-3,5a,6,7,8,9a-hexahydro-2-benzoxepine-4-carboxylic acid, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yan, Y, Zang, X, Cooper, S.J, Lin, H, Zhou, J, Tang, Y. | | Deposit date: | 2020-03-12 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.82449543 Å) | | Cite: | Biosynthesis of the fungal glyceraldehyde-3-phosphate dehydrogenase inhibitor heptelidic acid and mechanism of self-resistance
Chem Sci, 11, 2020
|
|
6YA6
 
 | | Minimal construct of Cdc7-Dbf4 bound to XL413 | | Descriptor: | 8-chloro-2-[(2S)-pyrrolidin-2-yl][1]benzofuro[3,2-d]pyrimidin-4(3H)-one, BORIC ACID, CHLORIDE ION, ... | | Authors: | Dick, S.D, Cherepanov, P. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis for the Activation and Target Site Specificity of CDC7 Kinase.
Structure, 28, 2020
|
|
6YA7
 
 | | Cdc7-Dbf4 bound to an Mcm2-S40 derived bivalent substrate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division cycle 7-related protein kinase,Cell division cycle 7-related protein kinase,Cell division cycle 7-related protein kinase, DNA replication licensing factor MCM2, ... | | Authors: | Dick, S.D, Cherepanov, P. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural Basis for the Activation and Target Site Specificity of CDC7 Kinase.
Structure, 28, 2020
|
|
6W4P
 
 | | CaMKII alpha-30 Cryo-EM reconstruction - Class B | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha | | Authors: | Chao, L.H, Stratton, M.M. | | Deposit date: | 2020-03-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Heterogeneity in human hippocampal CaMKII transcripts reveals allosteric hub-dependent regulation.
Sci.Signal., 13, 2020
|
|
