5QPQ
 
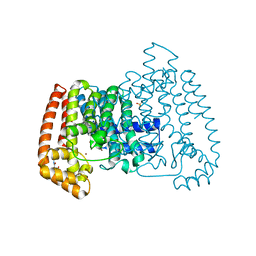 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOPL000631a | | Descriptor: | ACETATE ION, Farnesyl diphosphate synthase, SULFATE ION, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
5QQ8
 
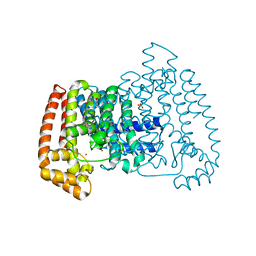 | | PanDDA analysis group deposition -- Crystal Structure of T. cruzi FPPS in complex with FMOOA000563a | | Descriptor: | 3-ethyl-1-[(1~{R},8~{S},9~{S},10~{S})-10-oxidanyl-11-oxatricyclo[6.2.1.0^{2,7}]undeca-2(7),3,5-trien-9-yl]imidazolidine-2,4-dione, ACETATE ION, Farnesyl diphosphate synthase, ... | | Authors: | Petrick, J.K, Nelson, E.R, Muenzker, L, Krojer, T, Douangamath, A, Brandao-Neto, J, von Delft, F, Dekker, C, Jahnke, W. | | Deposit date: | 2019-03-12 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PanDDA analysis group deposition - FPPS screened against the DSI Fragment Library
To Be Published
|
|
4N08
 
 | |
4DVH
 
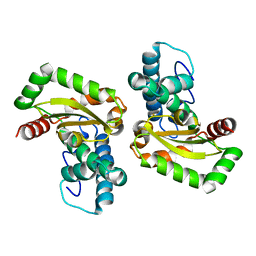 | | Crystal structure of Trypanosoma cruzi mitochondrial iron superoxide dismutase | | Descriptor: | FE (III) ION, Superoxide dismutase | | Authors: | Larrieux, N, Buschiazzo, A. | | Deposit date: | 2012-02-23 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and Molecular Basis of the Peroxynitrite-mediated Nitration and Inactivation of Trypanosoma cruzi Iron-Superoxide Dismutases (Fe-SODs) A and B: DISPARATE SUSCEPTIBILITIES DUE TO THE REPAIR OF TYR35 RADICAL BY CYS83 IN Fe-SODB THROUGH INTRAMOLECULAR ELECTRON TRANSFER.
J.Biol.Chem., 289, 2014
|
|
5V6V
 
 | | Crystal structure of small molecule aziridine 3 covalently bound to K-Ras G12C | | Descriptor: | 3-amino-1-{4-[6-chloro-8-fluoro-7-(5-methyl-1H-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, CALCIUM ION, GLYCEROL, ... | | Authors: | McGregor, L.M, Jenkins, M, Kerwin, C, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Expanding the Scope of Electrophiles Capable of Targeting K-Ras Oncogenes.
Biochemistry, 56, 2017
|
|
6CHP
 
 | |
6N9P
 
 | | Discovery of affinity-based probes for Btk occupancy assay | | Descriptor: | N-(3-{[2-amino-3-(4-phenoxyphenyl)pyridin-4-yl]oxy}phenyl)propanamide, Tyrosine-protein kinase BTK | | Authors: | Mochalkin, I. | | Deposit date: | 2018-12-03 | | Release date: | 2019-02-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery of Affinity-Based Probes for Btk Occupancy Assays.
ChemMedChem, 14, 2019
|
|
1YS3
 
 | |
5EQC
 
 | | Structure of the ornithine aminotransferase from Toxoplasma gondii crystallized in presence of oxidized glutathione reveals partial occupancy of PLP at the protein active site | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Minasov, G, Flores, K, Le, H.V, Silverman, R.B, McLeod, R.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-12 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the ornithine aminotransferase from Toxoplasma gondii crystallized in presence of oxidized glutathione reveals partial occupancy of PLP at the protein active site
To Be Published
|
|
7YY0
 
 | |
7YY1
 
 | |
7YY8
 
 | |
5NAH
 
 | |
6UVJ
 
 | |
5BSJ
 
 | |
5BS9
 
 | |
2G4R
 
 | | anomalous substructure of MogA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, molybdopterin biosynthesis Mog protein | | Authors: | Mueller-Dieckmann, C, Weiss, M.S. | | Deposit date: | 2006-02-22 | | Release date: | 2007-02-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | On the routine use of soft X-rays in macromolecular crystallography. Part IV. Efficient determination of anomalous substructures in biomacromolecules using longer X-ray wavelengths.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
5KAM
 
 | |
6BAI
 
 | | Multiconformer model of apo K197C PTP1B at 100 K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2017-10-12 | | Release date: | 2018-06-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
4FER
 
 | | Crystal structure of Bacillus Subtilis expansin (EXLX1) in complex with cellohexaose | | Descriptor: | ACETIC ACID, Expansin-yoaJ, GLYCEROL, ... | | Authors: | Georgelis, N, Yennawar, N.H, Cosgrove, D.J. | | Deposit date: | 2012-05-30 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Structural basis for entropy-driven cellulose binding by a type-A cellulose-binding module (CBM) and bacterial expansin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
6B95
 
 | | Multiconformer model of K197C PTP1B tethered to compound 2 at 100 K | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, N-(2',4'-difluoro-4-hydroxy[1,1'-biphenyl]-3-yl)-2-sulfanylacetamide, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2017-10-10 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
1RDP
 
 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV3 | | Descriptor: | 1,3-BIS-([[3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO-3,4-DIOXO-CYCLOBU TENYL]-AMINO-ETHYL]-AMINO-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TRIETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-05 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1RF2
 
 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV4 | | Descriptor: | 1,3-BIS-([3-[3-[3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYLAMINO-3,4-DIOXO-CYCLOBUTENYL]-AMINO-PROPOXY-ETHOXY-ETHOXY]-PROPYL-]AMINO-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, TRIETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-07 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
1RD9
 
 | | Cholera Toxin B-Pentamer Complexed With Bivalent Nitrophenol-Galactoside Ligand BV2 | | Descriptor: | 1,3-BIS-([3-(4-{3-[3-NITRO-5-(GALACTOPYRANOSYLOXY)-BENZOYLAMINO]-PROPYL}-PIPERAZIN-1-YL)-PROPYL-AMINO]-CARBONYLOXY)-2-AMINO-PROPANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HEXAETHYLENE GLYCOL, ... | | Authors: | Pickens, J.C, Mitchell, D.D, Liu, J, Tan, X, Zhang, Z, Verlinde, C.L, Hol, W.G, Fan, E. | | Deposit date: | 2003-11-05 | | Release date: | 2004-10-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Nonspanning bivalent ligands as improved surface receptor binding inhibitors of the cholera toxin B pentamer.
Chem.Biol., 11, 2004
|
|
7EDZ
 
 | | Crystal Structure of human PPCS in complex with P-HoPan and AMPPNP | | Descriptor: | 4-[[(2R)-3,3-dimethyl-2-oxidanyl-4-phosphonooxy-butanoyl]amino]butanoic acid, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sharma, N, Mostert, K.J, Strauss, E, Anand, R. | | Deposit date: | 2021-03-17 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Coenzyme A Level Modulator Hopantenate (HoPan) Inhibits Phosphopantotenoylcysteine Synthetase Activity.
Acs Chem.Biol., 16, 2021
|
|
