6L6V
 
 | | SPO1 Gp44 N-terminal region (1-55) | | Descriptor: | E3 protein | | Authors: | Liu, B, Wang, Z. | | Deposit date: | 2019-10-29 | | Release date: | 2021-05-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A Bacteriophage DNA Mimic Protein Employs a Non-specific Strategy to Inhibit the Bacterial RNA Polymerase.
Front Microbiol, 12, 2021
|
|
6JBX
 
 | | Crystal structure of Streptococcus pneumoniae FabT in complex with DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*GP*TP*TP*TP*GP*AP*CP*TP*GP*TP*CP*AP*AP*AP*TP*TP*AP*TP*G)-3'), DNA (5'-D(*CP*AP*TP*AP*AP*TP*TP*TP*GP*AP*CP*AP*GP*TP*CP*AP*AP*AP*CP*TP*AP*TP*T)-3'), Fatty acid biosynthesis transcriptional regulator, ... | | Authors: | Zuo, G, Chen, Z.P, Li, Q, Zhou, C.Z. | | Deposit date: | 2019-01-27 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into repression of the Pneumococcal fatty acid synthesis pathway by repressor FabT and co-repressor acyl-ACP.
Febs Lett., 593, 2019
|
|
4X8Q
 
 | | X-ray crystal structure of AlkD2 from Streptococcus mutans | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Mullins, E.A, Shi, R, Eichman, B.F. | | Deposit date: | 2014-12-10 | | Release date: | 2015-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.729 Å) | | Cite: | A New Family of HEAT-Like Repeat Proteins Lacking a Critical Substrate Recognition Motif Present in Related DNA Glycosylases.
Plos One, 10, 2015
|
|
4TR6
 
 | | Crystal structure of DNA polymerase sliding clamp from Bacillus subtilis | | Descriptor: | DNA polymerase III subunit beta, SODIUM ION | | Authors: | Burnouf, D, Olieric, V, Ennifar, E, Wolff, P. | | Deposit date: | 2014-06-14 | | Release date: | 2014-09-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Differential Modes of Peptide Binding onto Replicative Sliding Clamps from Various Bacterial Origins.
J.Med.Chem., 57, 2014
|
|
4UMK
 
 | | The complex of Spo0J and parS DNA in chromosomal partition system | | Descriptor: | DNA, PROBABLE CHROMOSOME-PARTITIONING PROTEIN PARB, SULFATE ION | | Authors: | Chen, B.W, Chu, C.H, Tung, J.Y, Hsu, C.E, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2014-05-19 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.096 Å) | | Cite: | Insights into ParB spreading from the complex structure of Spo0J and parS.
Proc. Natl. Acad. Sci. U.S.A., 112, 2015
|
|
5DDG
 
 | | The structure of transcriptional factor AraR from Bacteroides thetaiotaomicron VPI in complex with target double strand DNA | | Descriptor: | DNA (27-MER), FORMIC ACID, MALONIC ACID, ... | | Authors: | Chang, C, Tesar, C, Rodionov, D, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-08-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | A novel transcriptional regulator of L-arabinose utilization in human gut bacteria.
Nucleic Acids Res., 43, 2015
|
|
5YI3
 
 | | Structure of Lactococcus lactis ZitR, C30S mutant in complex with DNA | | Descriptor: | DNA (5'-D(*TP*GP*TP*TP*AP*AP*CP*TP*AP*GP*TP*TP*AP*AP*CP*A)-3'), ZINC ION, Zinc transport transcriptional regulator | | Authors: | Song, Y, Liu, H, Zhu, R, Yi, C, Chen, P. | | Deposit date: | 2017-10-01 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Allosteric histidine switch for regulation of intracellular zinc(II) fluctuation.
Proc.Natl.Acad.Sci.USA, 114, 2017
|
|
1LCD
 
 | | STRUCTURE OF THE COMPLEX OF LAC REPRESSOR HEADPIECE AND AN 11 BASE-PAIR HALF-OPERATOR DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), Lac Repressor, ... | | Authors: | Chuprina, V.P, Rullmann, J.A.C, Lamerichs, R.M.J.N, Van Boom, J.H, Boelens, R, Kaptein, R. | | Deposit date: | 1993-03-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 234, 1993
|
|
1LCC
 
 | | STRUCTURE OF THE COMPLEX OF LAC REPRESSOR HEADPIECE AND AN 11 BASE-PAIR HALF-OPERATOR DETERMINED BY NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | DNA (5'-D(*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*G)-3'), DNA (5'-D(*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*T)-3'), Lac Repressor, ... | | Authors: | Chuprina, V.P, Rullmann, J.A.C, Lamerichs, R.M.J.N, Van Boom, J.H, Boelens, R, Kaptein, R. | | Deposit date: | 1993-03-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the complex of lac repressor headpiece and an 11 base-pair half-operator determined by nuclear magnetic resonance spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 234, 1993
|
|
7VOX
 
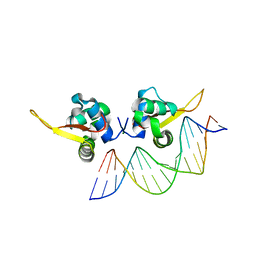 | | The crystal structure of human forkhead box protein A in complex with DNA 2 | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*AP*TP*TP*TP*AP*TP*TP*AP*TP*CP*GP*A)-3'), DNA (5'-D(P*TP*CP*GP*AP*TP*AP*AP*TP*AP*AP*AP*TP*AP*TP*TP*T)-3'), Hepatocyte nuclear factor 3-alpha, ... | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
7VOU
 
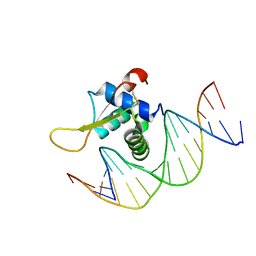 | | The crystal structure of human forkhead box protein in complex with DNA 1 | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*T)-3'), Forkhead box protein L2 | | Authors: | Choi, Y, Yoon, H.J, Lee, H.H. | | Deposit date: | 2021-10-14 | | Release date: | 2022-08-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | FOXL2 and FOXA1 cooperatively assemble on the TP53 promoter in alternative dimer configurations.
Nucleic Acids Res., 50, 2022
|
|
6FZS
 
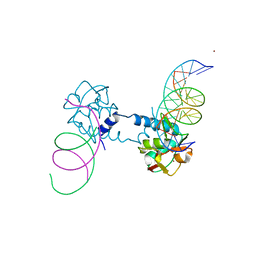 | | Crystal structure of Smad5-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*GP*GP*CP*GP*CP*GP*CP*CP*TP*GP*CP*A)-3'), Mothers against decapentaplegic homolog 5, ZINC ION | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2018-03-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
Computational and Structural Biotechnology Journal, 19, 2021
|
|
8IFO
 
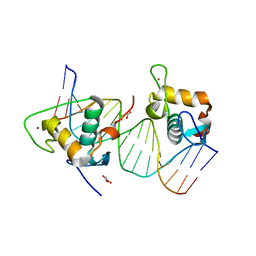 | | Crystal structure of estrogen related receptor-gamma DNA binding domain complexed with Pla2g12b promoter | | Descriptor: | DNA (5'-D(*GP*AP*GP*GP*AP*CP*AP*AP*AP*GP*GP*TP*GP*AP*AP*AP*C)-3'), DNA (5'-D(*GP*TP*TP*TP*CP*AP*CP*CP*TP*TP*TP*GP*TP*CP*CP*TP*C)-3'), Estrogen-related receptor gamma, ... | | Authors: | Xu, T, Zhen, X, Liu, J. | | Deposit date: | 2023-02-19 | | Release date: | 2023-03-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | ERR gamma-DBD undergoes dimerization and conformational rearrangement upon binding to the downstream site of the DR1 element.
Biochem.Biophys.Res.Commun., 656, 2023
|
|
6FZT
 
 | | Crystal structure of Smad8_9-MH1 bound to the GGCGC site. | | Descriptor: | DNA (5'-D(P*TP*GP*CP*AP*GP*GP*CP*GP*CP*GP*CP*CP*TP*GP*CP*A)-3'), GLYCEROL, Mothers against decapentaplegic homolog 9, ... | | Authors: | Kaczmarska, Z, Marquez, J.A, Macias, M.J. | | Deposit date: | 2018-03-15 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Unveiling the dimer/monomer propensities of Smad MH1-DNA complexes
Computational and Structural Biotechnology Journal, 19, 2021
|
|
8DVP
 
 | | Glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with substrate purine | | Descriptor: | ACETATE ION, Adenine DNA glycosylase, CALCIUM ION, ... | | Authors: | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2022-07-29 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
8DW0
 
 | | Glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with an enzyme-generated abasic site (AP) product and crystallized with sodium acetate | | Descriptor: | 1,2-ETHANEDIOL, Adenine DNA glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), ... | | Authors: | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2022-07-30 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
8DW4
 
 | | Glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with an abasic site product (AP) generated by the enzyme in crystals by removal of calcium | | Descriptor: | ACETATE ION, Adenine DNA glycosylase, DNA (5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3'), ... | | Authors: | Demir, M, Russelburg, L.P, Horvath, M.P, David, S.S. | | Deposit date: | 2022-07-31 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism.
Nucleic Acids Res., 51, 2023
|
|
8CSH
 
 | |
4G0U
 
 | | Human topoisomerase IIbeta in complex with DNA and amsacrine | | Descriptor: | DNA (5'-D(P*AP*GP*CP*CP*GP*AP*GP*C)-3'), DNA (5'-D(P*TP*GP*CP*AP*GP*CP*TP*CP*GP*GP*CP*T)-3'), DNA topoisomerase 2-beta, ... | | Authors: | Wu, C.C, Li, T.K, Li, Y.C, Chan, N.L. | | Deposit date: | 2012-07-10 | | Release date: | 2013-07-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | On the structural basis and design guidelines for type II topoisomerase-targeting anticancer drugs
Nucleic Acids Res., 41, 2013
|
|
2ZVK
 
 | | Crystal structure of PCNA in complex with DNA polymerase eta fragment | | Descriptor: | DNA polymerase eta, Proliferating cell nuclear antigen | | Authors: | Hishiki, A, Hashimoto, H, Hanafusa, T, Kamei, K, Ohashi, E, Shimizu, T, Ohmori, H, Sato, M. | | Deposit date: | 2008-11-11 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Basis for Novel Interactions between Human Translesion Synthesis Polymerases and Proliferating Cell Nuclear Antigen
J.Biol.Chem., 284, 2009
|
|
4WHV
 
 | | E3 ubiquitin-protein ligase RNF8 in complex with Ubiquitin-conjugating enzyme E2 N and Polyubiquitin-B | | Descriptor: | E3 ubiquitin-protein ligase RNF8, Polyubiquitin-B, Ubiquitin-conjugating enzyme E2 N, ... | | Authors: | Hodge, C.D, Edwards, R.A, Glover, J.N.M. | | Deposit date: | 2014-09-23 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (8.3 Å) | | Cite: | RNF8 E3 Ubiquitin Ligase Stimulates Ubc13 E2 Conjugating Activity That Is Essential for DNA Double Strand Break Signaling and BRCA1 Tumor Suppressor Recruitment.
J.Biol.Chem., 291, 2016
|
|
6X6E
 
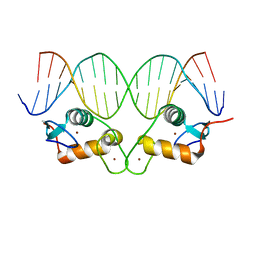 | |
8D4Y
 
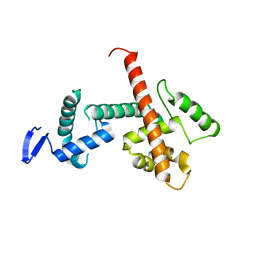 | | C-terminal SANT-SLIDE domain of human Chromodomain-helicase-DNA-binding protein 4 (CHD4) | | Descriptor: | Chromodomain-helicase-DNA-binding protein 4 | | Authors: | Moghaddas Sani, H, Deshpande, C.N, Panjikar, S, Patel, K, Mackay, J.P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-12-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The role of auxiliary domains in modulating CHD4 activity suggests mechanistic commonality between enzyme families.
Nat Commun, 13, 2022
|
|
6J4G
 
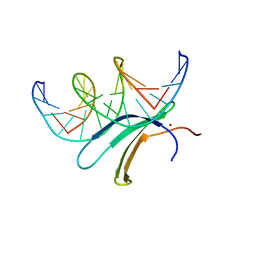 | | Crystal structure of the AtWRKY33 domain | | Descriptor: | DNA (5'-D(*AP*GP*CP*CP*TP*TP*TP*GP*AP*CP*CP*AP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*TP*GP*GP*TP*CP*AP*AP*AP*GP*GP*C)-3'), Probable WRKY transcription factor 33, ... | | Authors: | Xu, Y.P, Xu, H, Wang, B, Su, X.D. | | Deposit date: | 2019-01-08 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the N-terminal DNA binding domain of AtWRKY33
To Be Published
|
|
3E0C
 
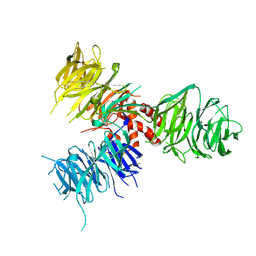 | | Crystal Structure of DNA Damage-Binding protein 1(DDB1) | | Descriptor: | DNA damage-binding protein 1 | | Authors: | Amaya, M.F, Xu, L, Hao, H, Bountra, C, Wickstroem, M, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structure and function of WD40 domain proteins.
Protein Cell, 2, 2011
|
|
