4RMH
 
 | | Human Sirt2 in complex with SirReal2 and Ac-Lys-H3 peptide | | Descriptor: | 2-[(4,6-dimethylpyrimidin-2-yl)sulfanyl]-N-[5-(naphthalen-1-ylmethyl)-1,3-thiazol-2-yl]acetamide, Ac-Lys-H3 peptide, NAD-dependent protein deacetylase sirtuin-2, ... | | Authors: | Rumpf, T, Schiedel, M, Karaman, B, Roessler, C, North, B.J, Lehotzky, A, Olah, J, Ladwein, K.I, Schmidtkunz, K, Gajer, M, Pannek, M, Steegborn, C, Sinclair, D.A, Gerhardt, S, Ovadi, J, Schutkowski, M, Sippl, W, Einsle, O, Jung, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Selective Sirt2 inhibition by ligand-induced rearrangement of the active site.
Nat Commun, 6, 2015
|
|
2K3Y
 
 | |
2L12
 
 | | Solution NMR structure of the chromobox protein 7 with H3K9me3 | | Descriptor: | Chromobox homolog 7, Histone H3 | | Authors: | Kaustov, L, Lemak, A, Gutmanas, A, Fares, C, Quang, H, Loppnau, P, Min, J, Edwards, A, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
2L11
 
 | | Solution NMR structure of the Cbx3 in complex with H3K9me3 peptide | | Descriptor: | Chromobox protein homolog 3, Histone H3 | | Authors: | Kaustov, L, Lemak, A, Fares, C, Gutmanas, A, Quang, H, Loppnau, P, Min, J, Edwards, A, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-22 | | Release date: | 2010-08-04 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
2L1B
 
 | | Solution NMR structure of the chromobox protein Cbx7 with H3K27me3 | | Descriptor: | Chromobox protein homolog 7, Histone H3 | | Authors: | Kaustov, L, Lemak, A, Fares, C, Gutmanas, A, Muhandiram, R, Quang, H, Loppnau, P, Min, J, Edwards, A, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-07-27 | | Release date: | 2010-08-25 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Recognition and specificity determinants of the human cbx chromodomains.
J.Biol.Chem., 286, 2011
|
|
2LGL
 
 | | NMR structure of the UHRF1 PHD domain | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, ZINC ION | | Authors: | Wang, C, Shen, J, Yang, Z, Chen, P, Zhao, B, Hu, W, Lan, W, Tong, X, Wu, H, Li, G, Cao, C. | | Deposit date: | 2011-07-28 | | Release date: | 2011-09-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for site-specific reading of unmodified R2 of histone H3 tail by UHRF1 PHD finger.
Cell Res., 21, 2011
|
|
8J71
 
 | |
8J70
 
 | | Native SAND domain of protein SP140 with DNA | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*GP*GP*CP*CP*GP*CP*CP*CP*T)-3'), Nuclear body protein SP140, selenourea | | Authors: | Li, H.T, Liu, W.Q. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Molecular basis for Speckled protein SP140 bivalent recognition of histone H3 and DNA.
To Be Published
|
|
1RJC
 
 | | Crystal structure of the camelid single domain antibody cAb-Lys2 in complex with hen egg white lysozyme | | Descriptor: | GLYCEROL, Lysozyme C, PHOSPHATE ION, ... | | Authors: | De Genst, E, Silence, K, Ghahroudi, M.A, Decanniere, K, Loris, R, Kinne, J, Wyns, L, Muyldermans, S. | | Deposit date: | 2003-11-19 | | Release date: | 2005-02-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Strong in vivo maturation compensates for structurally restricted H3 loops in antibody repertoires.
J.Biol.Chem., 280, 2005
|
|
1RI8
 
 | | Crystal Structure of the Camelid Single Domain Antibody 1D2L19 in complex with Hen Egg White Lysozyme | | Descriptor: | GLYCEROL, Lysozyme C, camelid ANTIBODY HEAVY CHAIN | | Authors: | De Genst, E, Silence, K, Ghahroudi, M.A, Decanniere, K, Loris, R, Kinne, J, Wyns, L, Muyldermans, S. | | Deposit date: | 2003-11-17 | | Release date: | 2005-02-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Strong in vivo maturation compensates for structurally restricted H3 loops in antibody repertoires.
J.Biol.Chem., 280, 2005
|
|
3OOI
 
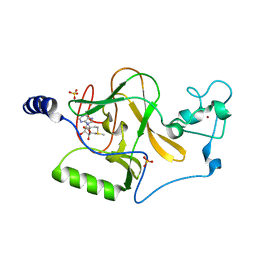 | | Crystal Structure of Human Histone-Lysine N-methyltransferase NSD1 SET domain in Complex with S-adenosyl-L-methionine | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-36 and H4 lysine-20 specific, S-ADENOSYLMETHIONINE, ... | | Authors: | Qiao, Q, Wang, M, Xu, R.M. | | Deposit date: | 2010-08-31 | | Release date: | 2010-12-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation
J.Biol.Chem., 286, 2010
|
|
1S55
 
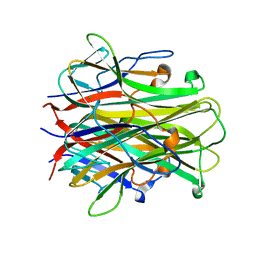 | | Mouse RANKL Structure at 1.9A Resolution | | Descriptor: | CHLORIDE ION, Tumor necrosis factor ligand superfamily member 11 | | Authors: | Teale, M.J, Feug, X, Chen, L, Bice, T, Meehan, E.J. | | Deposit date: | 2004-01-19 | | Release date: | 2005-07-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Murine RANKL Extra Cellular Domain Homotrimer Structure In Space Groups P212121 And H3 At 1.9 And 2.6 Respectively
To be Published
|
|
7W6L
 
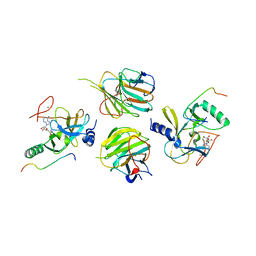 | | The crystal structure of MLL3-RBBP5-ASH2L in complex with H3K4me0 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2C, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W6I
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me1 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
7W67
 
 | | The crystal structure of MLL1 (N3861I/Q3867L/C3882SS)-RBBP5-ASH2L in complex with H3K4me0 peptide | | Descriptor: | Histone H3.3C, Histone-lysine N-methyltransferase 2A, Retinoblastoma-binding protein 5, ... | | Authors: | Zhao, L, Li, Y, Chen, Y. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural basis for product specificities of MLL family methyltransferases.
Mol.Cell, 82, 2022
|
|
6OCB
 
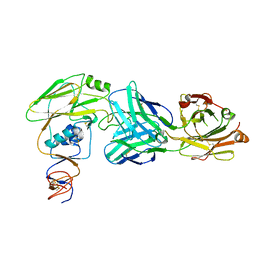 | |
7VYW
 
 | |
7VZ2
 
 | | Crystal structure of chromodomain of Arabidopsis LHP1 | | Descriptor: | Chromo domain-containing protein LHP1, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Min, J. | | Deposit date: | 2021-11-15 | | Release date: | 2022-02-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the recognition of methylated histone H3 by the Arabidopsis LHP1 chromodomain.
J.Biol.Chem., 298, 2022
|
|
6VDB
 
 | | SETD2 in complex with a H3-variant super-substrate peptide | | Descriptor: | ALA-PRO-ARG-PHE-GLY-GLY-VAL-MET-ARG-PRO-ASN-ARG, Histone-lysine N-methyltransferase SETD2, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Beldar, S, Tempel, W, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Jeltsch, A, Min, J, Structural Genomics Consortium, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-12-24 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Sequence specificity analysis of the SETD2 protein lysine methyltransferase and discovery of a SETD2 super-substrate.
Commun Biol, 3, 2020
|
|
6VIL
 
 | |
7XI9
 
 | | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-ordered form) | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*TP*CP*(C55)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Onoda, H, Kikuchi, A, Kori, S, Yoshimi, S, Yamagata, A, Arita, K. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Structural basis for activation of DNMT1.
Nat Commun, 13, 2022
|
|
7XIB
 
 | | Cryo-EM structure of human DNMT1 (aa:351-1616) in complex with ubiquitinated H3 and hemimethylated DNA analog (CXXC-disordered form) | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*AP*(5CM)P*GP*GP*AP*AP*GP*G)-3'), DNA (5'-D(*CP*CP*TP*TP*CP*(C55)P*GP*TP*AP*AP*GP*T)-3'), DNA (cytosine-5)-methyltransferase 1, ... | | Authors: | Onoda, H, Kikuchi, A, Kori, S, Yoshimi, S, Yamagata, A, Arita, K. | | Deposit date: | 2022-04-12 | | Release date: | 2022-11-30 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Structural basis for activation of DNMT1.
Nat Commun, 13, 2022
|
|
4X8N
 
 | | Crystal structure of Ash2L SPRY domain in complex with phosphorylated RbBP5 | | Descriptor: | Retinoblastoma-binding protein 5, Set1/Ash2 histone methyltransferase complex subunit ASH2 | | Authors: | Zhang, P, Chaturvedi, C.P, Brunzelle, J.S, Skiniotis, G, Brand, M, Shilatifard, A, Couture, J.-F. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A phosphorylation switch on RbBP5 regulates histone H3 Lys4 methylation.
Genes Dev., 29, 2015
|
|
4X8P
 
 | | Crystal structure of Ash2L SPRY domain in complex with RbBP5 | | Descriptor: | GLYCEROL, Retinoblastoma-binding protein 5, Set1/Ash2 histone methyltransferase complex subunit ASH2,Set1/Ash2 histone methyltransferase complex subunit ASH2 | | Authors: | Zhang, P, Chaturvedi, C.P, Brunzelle, J.S, Skiniotis, G, Brand, M, Shilatifard, A, Couture, J.-F. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A phosphorylation switch on RbBP5 regulates histone H3 Lys4 methylation.
Genes Dev., 29, 2015
|
|
7E7A
 
 | | Crystal structure of apo ENL YEATS domain T3 mutant | | Descriptor: | Protein ENL | | Authors: | Li, Y, Li, H. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Crystal structure of ENL YEATS domain T1 mutant in complex with histone H3 acetylation at K27
To Be Published
|
|
