8XKY
 
 | | Structure of the TOM40 complex annealed | | Descriptor: | Mitochondrial import receptor subunit TOM22, Mitochondrial import receptor subunit TOM40, Mitochondrial import receptor subunit TOM5, ... | | Authors: | Yang, L.Y, Shen, Q.T. | | Deposit date: | 2023-12-25 | | Release date: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structure of the TOM40 complex annealed
To Be Published
|
|
5IEE
 
 | | Murine endoplasmic reticulum alpha-glucosidase II with 1-deoxynojirimycin | | Descriptor: | 1,2-ETHANEDIOL, 1-DEOXYNOJIRIMYCIN, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-02-25 | | Release date: | 2016-07-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5LY7
 
 | | Crystal structure of NagZ H174A mutant from Pseudomonas aeruginosa in complex with the inhibitor 2-acetamido-1,2-dideoxynojirimycin | | Descriptor: | 2-ACETAMIDO-1,2-DIDEOXYNOJIRMYCIN, Beta-hexosaminidase, DI(HYDROXYETHYL)ETHER | | Authors: | Acebron, I, Artola-Recolons, C, Mahasenan, K, Mobashery, S, Hermoso, J.A. | | Deposit date: | 2016-09-25 | | Release date: | 2017-05-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Catalytic Cycle of the N-Acetylglucosaminidase NagZ from Pseudomonas aeruginosa.
J. Am. Chem. Soc., 139, 2017
|
|
5NVR
 
 | |
6R9I
 
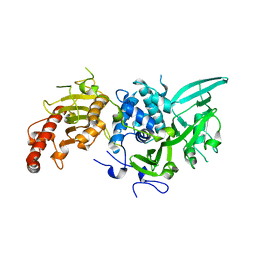 | |
5CYZ
 
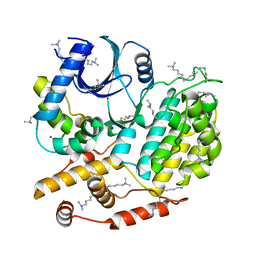 | | Structure of S. cerevisiae Hrr25:Mam1 complex, form 1 | | Descriptor: | Casein kinase I homolog HRR25, Monopolin complex subunit MAM1, ZINC ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
5EL6
 
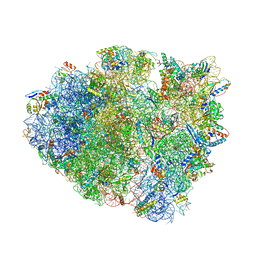 | | Structure of T. thermophilus 70S ribosome complex with mRNA and tRNALys in the A-site with a U-U mismatch in the first position and antibiotic paromomycin | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Rozov, A, Demeshkina, N, Khusainov, I, Yusupov, M, Yusupova, G. | | Deposit date: | 2015-11-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code.
Nat Commun, 7, 2016
|
|
8A2C
 
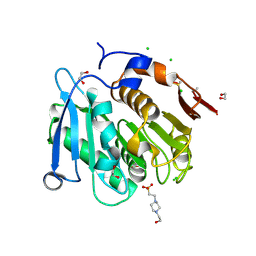 | | The crystal structure of the S178A mutant of PET40, a PETase enzyme from an unclassified Amycolatopsis | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Costanzi, E, Applegate, V, Port, A, Smits, S.H.J. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The metagenome-derived esterase PET40 is highly promiscuous and hydrolyses polyethylene terephthalate (PET).
Febs J., 291, 2024
|
|
5IWU
 
 | |
8ZPW
 
 | |
5CZO
 
 | | Structure of S. cerevisiae Hrr25:Mam1 complex, form 2 | | Descriptor: | Casein kinase I homolog HRR25, Monopolin complex subunit MAM1, ZINC ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-07-31 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
1DS3
 
 | | CRYSTAL STRUCTURE OF OMTKY3-CH2-ASP19I | | Descriptor: | OVOMUCOID | | Authors: | Bateman, K.S, Huang, K, Anderson, S, Lu, W, Qasim, M.A, Laskowski Jr, M, James, M.N.G. | | Deposit date: | 2000-01-06 | | Release date: | 2001-01-31 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Contribution of peptide bonds to inhibitor-protease binding: crystal structures of the turkey ovomucoid third domain backbone variants OMTKY3-Pro18I and OMTKY3-psi[COO]-Leu18I in complex with Streptomyces griseus proteinase B (SGPB) and the structure of the free inhibitor, OMTKY-3-psi[CH2NH2+]-Asp19I
J.Mol.Biol., 305, 2001
|
|
6CBO
 
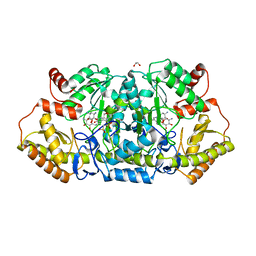 | | X-ray structure of GenB1 from micromonospora echinospora in complex with neamine and PLP (as the external aldimine) | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl 2-amino-2,6-dideoxy-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]-alpha-D-glucopyranoside, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Dow, G.T, Thoden, J.B, Holden, H.M. | | Deposit date: | 2018-02-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The three-dimensional structure of NeoB: An aminotransferase involved in the biosynthesis of neomycin.
Protein Sci., 27, 2018
|
|
4URO
 
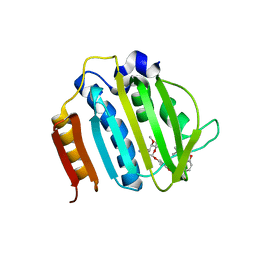 | | Crystal Structure of Staph GyraseB 24kDa in complex with Novobiocin | | Descriptor: | DNA GYRASE SUBUNIT B, NOVOBIOCIN | | Authors: | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | Deposit date: | 2014-07-01 | | Release date: | 2014-07-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
6CFD
 
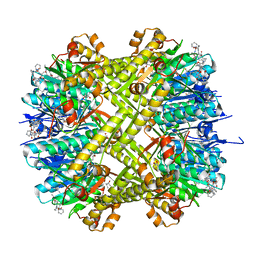 | | ADEP4 bound to E. faecium ClpP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ATP-dependent Clp protease proteolytic subunit, N-[(6aS,12S,15aS,17R,21R,23aS)-17,21-dimethyl-6,11,15,20,23-pentaoxooctadecahydro-2H,6H,11H,15H-pyrido[2,1-i]dipyrrolo[2,1-c:2',1'-l][1,4,7,10,13]oxatetraazacyclohexadecin-12-yl]-3,5-difluoro-Nalpha-[(2E)-hept-2-enoyl]-L-phenylalaninamide | | Authors: | Lee, R.E, Griffith, E.C. | | Deposit date: | 2018-02-14 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | In VivoandIn VitroEffects of a ClpP-Activating Antibiotic against Vancomycin-Resistant Enterococci.
Antimicrob. Agents Chemother., 62, 2018
|
|
5H29
 
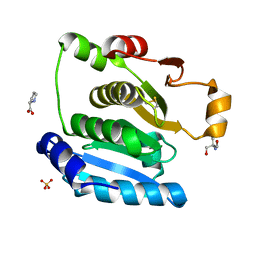 | | Crystal Structure of the NTD_N/C domain of Alkylhydroperoxide Reductase AhpF from Enterococcus Faecalis (V583) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PROLINE, SULFATE ION, ... | | Authors: | Balakrishna, A.M, Kwang, T.Y, Gruber, G. | | Deposit date: | 2016-10-14 | | Release date: | 2017-11-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel insights into the vancomycin-resistant Enterococcus faecalis (V583) alkylhydroperoxide reductase subunit F
Biochim. Biophys. Acta, 1861, 2017
|
|
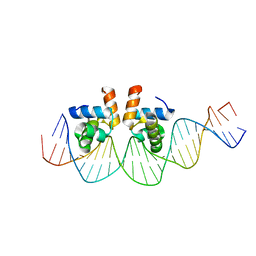
4WUH
 
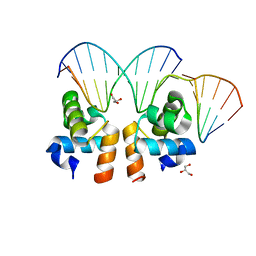 | | Crystal structure of E. faecalis DNA binding domain LiaR wild type complexed with 22bp DNA | | Descriptor: | DNA (5'-D(P*AP*AP*AP*TP*CP*G)-3'), DNA (5'-D(P*GP*GP*AP*CP*TP*TP*AP*AP*GP*AP*AP*CP*GP*AP*TP*TP*T)-3'), DNA (5'-D(P*TP*TP*CP*TP*TP*AP*AP*GP*TP*CP*C)-3'), ... | | Authors: | Davlieva, M, Shamoo, Y. | | Deposit date: | 2014-10-31 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | A variable DNA recognition site organization establishes the LiaR-mediated cell envelope stress response of enterococci to daptomycin.
Nucleic Acids Res., 43, 2015
|
|
7AQS
 
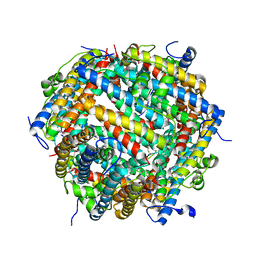 | | Crystal structure of E. coli DPS in space group P1 | | Descriptor: | DNA protection during starvation protein, FE (III) ION | | Authors: | Jakob, R.P, Pipercevic, J, Righetto, R, Goldie, K, Stahlberg, H, Maier, T, Hiller, S. | | Deposit date: | 2020-10-22 | | Release date: | 2021-09-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of a Dps contamination in Mitomycin-C-induced expression of Colicin Ia.
Biochim Biophys Acta Biomembr, 1863, 2021
|
|
4WSZ
 
 | |
8QX8
 
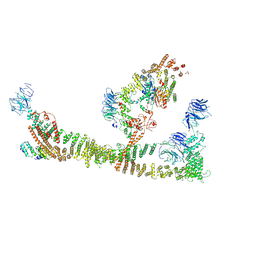 | | Endosomal membrane tethering complex CORVET | | Descriptor: | E3 ubiquitin-protein ligase PEP5, Vacuolar membrane protein PEP3, Vacuolar protein sorting-associated protein 16, ... | | Authors: | Shvarev, D, Ungermann, C, Moeller, A. | | Deposit date: | 2023-10-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the endosomal CORVET tethering complex.
Nat Commun, 15, 2024
|
|
8QOF
 
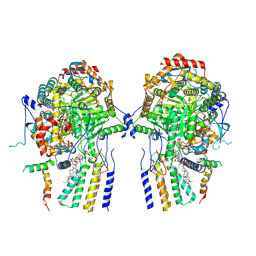 | | Cryo-EM structure of the yeast SPT-Orm2-Dimer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ORM2 isoform 1, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Schaefer, J, Koerner, C, Moeller, A, Froehlich, F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of the Orm2-containing serine palmitoyltransferase complex reveals distinct inhibitory potentials of yeast Orm proteins.
Cell Rep, 43, 2024
|
|
8QOG
 
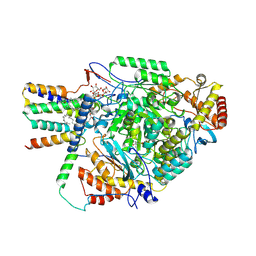 | | Cryo-EM structure of the yeast SPT-Orm2-Monomer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ORM2 isoform 1, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Schaefer, J, Koerner, C, Moeller, A, Froehlich, F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The structure of the Orm2-containing serine palmitoyltransferase complex reveals distinct inhibitory potentials of yeast Orm proteins.
Cell Rep, 43, 2024
|
|
7YHE
 
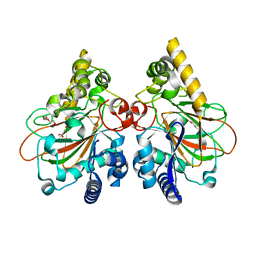 | | Crystal structure of the triple mutant CmnC-L136Q,S138G,D249Y in complex with alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, CmnC, FE (III) ION, ... | | Authors: | Huang, S.J, Hsiao, Y.H, Lin, E.C, Hsiao, P.Y, Chang, C.Y. | | Deposit date: | 2022-07-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Crystal structure of the alpha-ketoglutarate-dependent non-heme iron oxygenase CmnC in capreomycin biosynthesis and its engineering to catalyze hydroxylation of the substrate enantiomer.
Front Chem, 10, 2022
|
|
8QTN
 
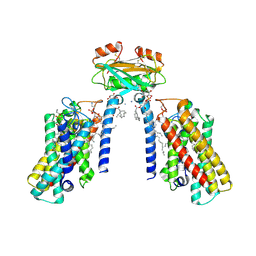 | | Cryo-EM structure of the apo yeast Ceramide Synthase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, AMMONIUM ION, Ceramide synthase LAC1, ... | | Authors: | Schaefer, J, Clausmeyer, L, Koerner, C, Moeller, A, Froehlich, F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the yeast ceramide synthase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
4WUL
 
 | |
