8WG0
 
 | | Crystal structure of GH97 glucodextranase from Flavobacterium johnsoniae in complex with glucose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Candidate alpha-glucosidase Glycoside hydrolase family 97, ... | | Authors: | Kurata, R, Nakamura, S, Miyazaki, T. | | Deposit date: | 2023-09-20 | | Release date: | 2024-05-08 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into alpha-(1→6)-linkage preference of GH97 glucodextranase from Flavobacterium johnsoniae.
Febs J., 291, 2024
|
|
8WGT
 
 | | Crystal structure of V30M-TTR in complex with compound 7 | | Descriptor: | Transthyretin, [4,7-bis(chloranyl)-2-ethyl-1-benzofuran-3-yl]-[3,5-bis(iodanyl)-4-oxidanyl-phenyl]methanone | | Authors: | Yokoyama, T. | | Deposit date: | 2023-09-22 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Development of Benziodarone Analogues with Enhanced Potency for Selective Binding to Transthyretin in Human Plasma.
J.Med.Chem., 67, 2024
|
|
7RQE
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site deacylated tRNA analog CACCA, P-site MAI-tripeptidyl-tRNA analog ACCA-IAM, and chloramphenicol at 2.40A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RQA
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MTI-tripeptidyl-tRNA analog ACCA-ITM at 2.40A resolution | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PZY
 
 | | Structure of the vacant Candida albicans 80S ribosome | | Descriptor: | 1,4-DIAMINOBUTANE, 18S ribosomal RNA, 25S ribosomal RNA, ... | | Authors: | Zgadzay, Y, Kolosova, O, Stetsenko, A, Jenner, L, Guskov, A, Yusupova, G, Yusupov, M. | | Deposit date: | 2021-10-13 | | Release date: | 2022-05-18 | | Last modified: | 2022-06-08 | | Method: | ELECTRON MICROSCOPY (2.32 Å) | | Cite: | E-site drug specificity of the human pathogen Candida albicans ribosome.
Sci Adv, 8, 2022
|
|
8VTY
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with ciprofloxacin and protein Y at 2.60A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-CYCLOPROPYL-6-FLUORO-4-OXO-7-PIPERAZIN-1-YL-1,4-DIHYDROQUINOLINE-3-CARBOXYLIC ACID, 16S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 20, 2024
|
|
7P81
 
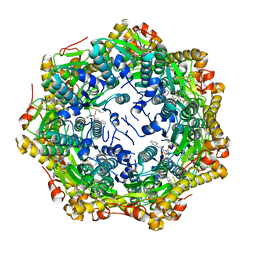 | | Crystal structure of ClpP from Bacillus subtilis in complex with ADEP2 (compact state) | | Descriptor: | ADEP2, ATP-dependent Clp protease proteolytic subunit | | Authors: | Lee, B.-G, Kim, L, Kim, M.K, Kwon, D.H, Song, H.K. | | Deposit date: | 2021-07-21 | | Release date: | 2022-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural insights into ClpP protease side exit pore-opening by a pH drop coupled with substrate hydrolysis.
Embo J., 41, 2022
|
|
7QIZ
 
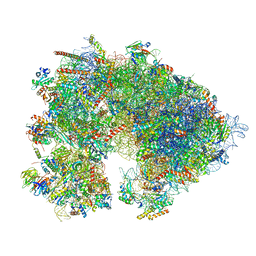 | | Specific features and methylation sites of a plant 80S ribosome | | Descriptor: | 1,4-DIAMINOBUTANE, 18S, 25S rRNA, ... | | Authors: | Cottilli, P, Itoh, Y, Amunts, A. | | Deposit date: | 2021-12-16 | | Release date: | 2022-08-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Cryo-EM structure and rRNA modification sites of a plant ribosome.
Plant Commun., 3, 2022
|
|
8VTU
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolone MCX-66, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.40A resolution | | Descriptor: | 1-cyclopropyl-7-[(4-{[3-({(3aR,4R,7R,8S,9S,10R,11R,13R,14E,15S,15aR)-10-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy}-4-ethyl-11-methoxy-3a,7,9,11,13,15-hexamethyl-14-[({3-[5-(methylcarbamoyl)pyridin-3-yl]prop-2-yn-1-yl}oxy)imino]-2,6-dioxododecahydro-2H,4H-[1,3]dioxolo[4,5-c]oxacyclotetradecin-8-yl}oxy)-3-oxopropyl]amino}butyl)amino]-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 20, 2024
|
|
8VTV
 
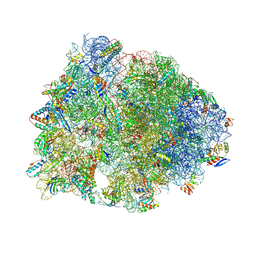 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolone MCX-91, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.55A resolution | | Descriptor: | 1-cyclopropyl-7-{7-[({[(3aR,4R,7R,8S,9S,10R,11R,13R,14E,15S,15aR)-10-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy}-4-ethyl-11-methoxy-14-(methoxyimino)-3a,7,9,11,13,15-hexamethyl-2,6-dioxododecahydro-2H,4H-[1,3]dioxolo[4,5-c]oxacyclotetradecin-8-yl]oxy}carbonyl)amino]hept-1-yn-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 20, 2024
|
|
8VTX
 
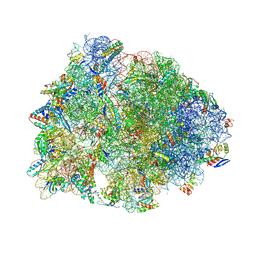 | | Crystal structure of the A2058-N6-dimethylated Thermus thermophilus 70S ribosome in complex with macrolone MCX-128, mRNA, aminoacylated A-site Phe-tRNAphe, aminoacylated P-site fMet-tRNAmet, and deacylated E-site tRNAphe at 2.40A resolution | | Descriptor: | 1-cyclopropyl-7-(4-{4-[({[(3aR,4R,7R,8S,9S,10R,11R,13R,14E,15S,15aR)-10-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy}-4-ethyl-11-methoxy-14-(methoxyimino)-3a,7,9,11,13,15-hexamethyl-2,6-dioxododecahydro-2H,4H-[1,3]dioxolo[4,5-c]oxacyclotetradecin-8-yl]oxy}carbonyl)amino]butyl}piperazin-1-yl)-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (non-preferred name), 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 20, 2024
|
|
8VTW
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with macrolone MCX-128 and protein Y at 2.35A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-cyclopropyl-7-(4-{4-[({[(3aR,4R,7R,8S,9S,10R,11R,13R,14E,15S,15aR)-10-{[(2S,3R,4S,6R)-4-(dimethylamino)-3-hydroxy-6-methyloxan-2-yl]oxy}-4-ethyl-11-methoxy-14-(methoxyimino)-3a,7,9,11,13,15-hexamethyl-2,6-dioxododecahydro-2H,4H-[1,3]dioxolo[4,5-c]oxacyclotetradecin-8-yl]oxy}carbonyl)amino]butyl}piperazin-1-yl)-6-fluoro-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (non-preferred name), 16S Ribosomal RNA, ... | | Authors: | Aleksandrova, E.V, Ma, C.-X, Klepacki, D, Alizadeh, F, Vazquez-Laslop, N, Liang, J.-H, Polikanov, Y.S, Mankin, A.S. | | Deposit date: | 2024-01-27 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Macrolones target bacterial ribosomes and DNA gyrase and can evade resistance mechanisms.
Nat.Chem.Biol., 20, 2024
|
|
1SNT
 
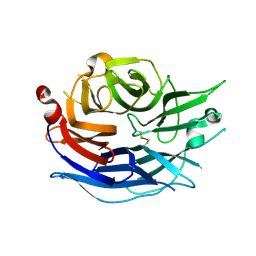 | | Structure of the human cytosolic sialidase Neu2 | | Descriptor: | Sialidase 2 | | Authors: | Chavas, L.M.G, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Kato, R, Monti, E, Wakatsuki, S. | | Deposit date: | 2004-03-12 | | Release date: | 2004-11-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Human Cytosolic Sialidase Neu2: EVIDENCE FOR THE DYNAMIC NATURE OF SUBSTRATE RECOGNITION
J.Biol.Chem., 280, 2005
|
|
8W4Y
 
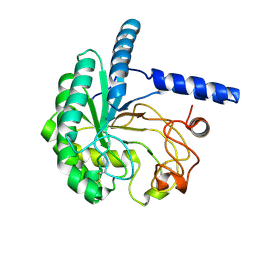 | | Neutron structure of cellulase Cel6A from Phanerochaete chrysosporium at room temperature, low-D2O-solvent | | Descriptor: | Glucanase | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-08-25 | | Release date: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.4 Å), X-RAY DIFFRACTION | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
8W8U
 
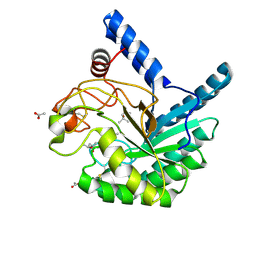 | | High-resolution X-ray structure of cellulase Cel6A from Phanerochaete chrysosporium at cryogenic temperature | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, Glucanase | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-09-04 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
8W4X
 
 | | Neutron structure of cellulase Cel6A from Phanerochaete chrysosporium at room temperature, Enzyme-Product complex | | Descriptor: | Glucanase, SODIUM ION, beta-D-glucopyranose, ... | | Authors: | Tachioka, M, Yamaguchi, S, Nakamura, A, Ishida, T, Kusaka, K, Yamada, T, Yano, N, Chatake, T, Tamada, T, Takeda, K, Niwa, S, Tanaka, H, Takahashi, S, Inaka, K, Furubayashi, N, Deguchi, S, Samejima, M, Igarashi, K. | | Deposit date: | 2023-08-25 | | Release date: | 2025-03-12 | | Method: | NEUTRON DIFFRACTION (1.4 Å), X-RAY DIFFRACTION | | Cite: | Deprotonated Arginine Controls a Putative Catalytic Base in Invert-ing Family 6 Glycoside Hydrolase
To Be Published
|
|
8WL5
 
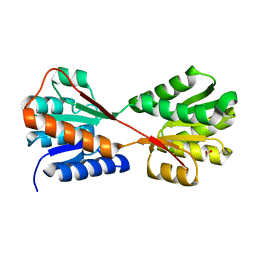 | | X-ray structure of Enterobacter cloacae allose-binding protein in free form | | Descriptor: | 1,2-ETHANEDIOL, Allose ABC transporter | | Authors: | Kamitori, S. | | Deposit date: | 2023-09-29 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | X-ray structures of Enterobacter cloacae allose-binding protein in complexes with monosaccharides demonstrate its unique recognition mechanism for high affinity to allose.
Biochem.Biophys.Res.Commun., 682, 2023
|
|
1SO7
 
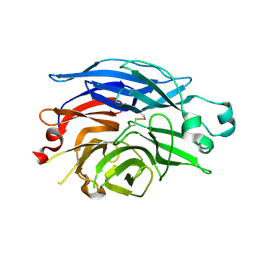 | | Maltose-induced structure of the human cytolsolic sialidase Neu2 | | Descriptor: | CHLORIDE ION, Sialidase 2 | | Authors: | Chavas, L.M.G, Fusi, P, Tringali, C, Venerando, B, Tettamanti, G, Kato, R, Monti, E, Wakatsuki, S. | | Deposit date: | 2004-03-12 | | Release date: | 2004-11-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal Structure of the Human Cytosolic Sialidase Neu2: EVIDENCE FOR THE DYNAMIC NATURE OF SUBSTRATE RECOGNITION
J.Biol.Chem., 280, 2005
|
|
7RQC
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MFI-tripeptidyl-tRNA analog ACCA-IFM at 2.50A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RQB
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with protein Y, A-site aminoacyl-tRNA analog ACC-PMN, and P-site MAI-tripeptidyl-tRNA analog ACCA-IAM at 2.45A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Syroegin, E.A, Flemmich, L, Klepacki, D, Vazquez-Laslop, N, Micura, R, Polikanov, Y.S. | | Deposit date: | 2021-08-06 | | Release date: | 2022-01-26 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural basis for the context-specific action of the classic peptidyl transferase inhibitor chloramphenicol.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7UG7
 
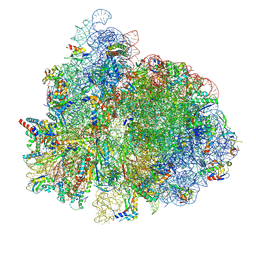 | | 70S ribosome complex in an intermediate state of translocation bound to EF-G(GDP) stalled by Argyrin B | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Rundlet, E.J, Wieland, M, Holm, M, Koller, T.O, Blanchard, S.C, Wilson, D.N. | | Deposit date: | 2022-03-24 | | Release date: | 2022-05-18 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | The cyclic octapeptide antibiotic argyrin B inhibits translation by trapping EF-G on the ribosome during translocation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UNW
 
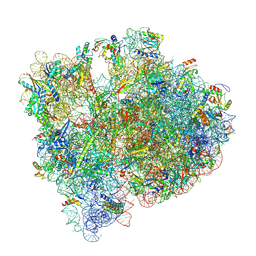 | | Pseudomonas aeruginosa 70S ribosome initiation complex bound to IF2-GDPCP (structure II-B) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Basu, R.S, Sherman, M.B, Gagnon, M.G. | | Deposit date: | 2022-04-11 | | Release date: | 2022-06-22 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Compact IF2 allows initiator tRNA accommodation into the P site and gates the ribosome to elongation
Nat Commun, 13, 2022
|
|
7UNV
 
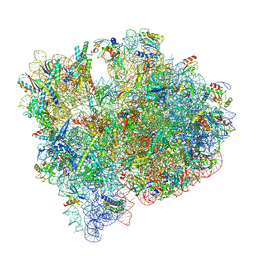 | | Pseudomonas aeruginosa 70S ribosome initiation complex bound to IF2-GDPCP (structure II-A) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Basu, R.S, Sherman, M.B, Gagnon, M.G. | | Deposit date: | 2022-04-11 | | Release date: | 2022-06-22 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Compact IF2 allows initiator tRNA accommodation into the P site and gates the ribosome to elongation
Nat Commun, 13, 2022
|
|
7UNU
 
 | | Pseudomonas aeruginosa 70S ribosome initiation complex bound to compact IF2-GDP (composite structure I-B) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Basu, R.S, Sherman, M.B, Gagnon, M.G. | | Deposit date: | 2022-04-11 | | Release date: | 2022-06-22 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Compact IF2 allows initiator tRNA accommodation into the P site and gates the ribosome to elongation
Nat Commun, 13, 2022
|
|
7UNR
 
 | | Pseudomonas aeruginosa 70S ribosome initiation complex bound to compact IF2-GDP (composite structure I-A) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Basu, R.S, Sherman, M.B, Gagnon, M.G. | | Deposit date: | 2022-04-11 | | Release date: | 2022-06-22 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Compact IF2 allows initiator tRNA accommodation into the P site and gates the ribosome to elongation
Nat Commun, 13, 2022
|
|
