8U81
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State A From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U83
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State C From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.975 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U84
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State D From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8VEX
 
 | |
8VEY
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and TNG908 | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, CHLORIDE ION, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
8VET
 
 | |
8VEU
 
 | |
8VEW
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA and oxamide compound 24 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, 5-{2-[(2R,5S)-5-methyl-2-phenylpiperidin-1-yl](oxo)acetamido}pyridine-3-carboxamide, CHLORIDE ION, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
8VEO
 
 | | Crystal structure of PRMT5:MEP50 in complex with MTA | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXY-5'-METHYLTHIOADENOSINE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Whittington, D.A. | | Deposit date: | 2023-12-20 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery of TNG908: A Selective, Brain Penetrant, MTA-Cooperative PRMT5 Inhibitor That Is Synthetically Lethal with MTAP -Deleted Cancers.
J.Med.Chem., 67, 2024
|
|
8URQ
 
 | | Spo11 core complex with gapped DNA | | Descriptor: | Antiviral protein SKI8, MAGNESIUM ION, Meiosis-specific protein SPO11, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the yeast Spo11 core complex bound to DNA
To Be Published
|
|
8WQR
 
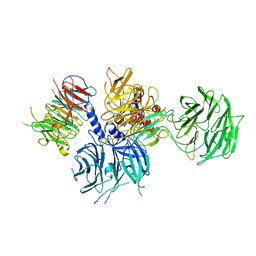 | | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation | | Descriptor: | Activating molecule in BECN1-regulated autophagy protein 1, DNA damage-binding protein 1 | | Authors: | Liu, M, Wang, Y, Su, M.Y, Stjepanovic, G. | | Deposit date: | 2023-10-12 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structure of the DDB1-AMBRA1 E3 ligase receptor complex linked to cell cycle regulation.
Nat Commun, 14, 2023
|
|
8VY7
 
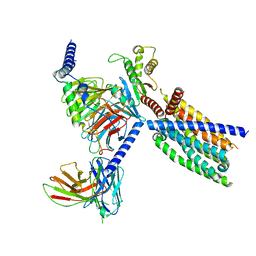 | | CryoEM structure of Gi-coupled TAS2R14 with cholesterol and an intracellular tastant | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kim, Y, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2024-02-07 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Bitter taste receptor activation by cholesterol and an intracellular tastant.
Nature, 628, 2024
|
|
8VY9
 
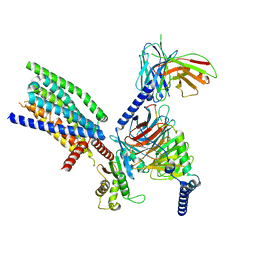 | | CryoEM structure of Ggust-coupled TAS2R14 with cholesterol and an intracellular tastant | | Descriptor: | 4-methyl-N-[(2M)-2-(1H-tetrazol-5-yl)phenyl]-6-(trifluoromethyl)pyrimidin-2-amine, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kim, Y, Gumpper, R.H, Roth, B.L. | | Deposit date: | 2024-02-07 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Bitter taste receptor activation by cholesterol and an intracellular tastant.
Nature, 628, 2024
|
|
8URU
 
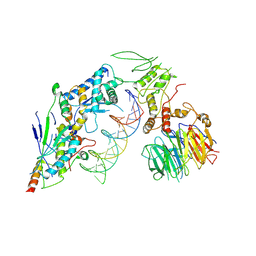 | | Spo11 core complex with hairpin DNA | | Descriptor: | Antiviral protein SKI8, Hairpin DNA, MAGNESIUM ION, ... | | Authors: | Yu, Y, Patel, D.J. | | Deposit date: | 2023-10-26 | | Release date: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of the yeast Spo11 core complex bound to DNA
To Be Published
|
|
7VPG
 
 | | Crystal structure of the C-terminal tail of SARS-CoV-1 Orf6 complex with human nucleoporin pair Rae1-Nup98 | | Descriptor: | Isoform 3 of Nuclear pore complex protein Nup98-Nup96, ORF6 protein, mRNA export factor | | Authors: | Li, T, Guo, H, Yang, T, Wen, Y, Ji, X. | | Deposit date: | 2021-10-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Molecular Mechanism of SARS-CoVs Orf6 Targeting the Rae1-Nup98 Complex to Compete With mRNA Nuclear Export.
Front Mol Biosci, 8, 2021
|
|
7VPH
 
 | | Crystal structure of the C-terminal tail of SARS-CoV-2 Orf6 complex with human nucleoporin pair Rae1-Nup98 | | Descriptor: | Isoform 3 of Nuclear pore complex protein Nup98-Nup96, ORF6 protein, mRNA export factor | | Authors: | Li, T, Guo, H, Yang, T, Wen, Y, Ji, X. | | Deposit date: | 2021-10-17 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular Mechanism of SARS-CoVs Orf6 Targeting the Rae1-Nup98 Complex to Compete With mRNA Nuclear Export.
Front Mol Biosci, 8, 2021
|
|
7VOP
 
 | | Cryo-EM structure of Xenopus laevis nuclear pore complex cytoplasmic ring subunit | | Descriptor: | GATOR complex protein SEC13, IL4I1 protein, MGC154553 protein, ... | | Authors: | Tai, L, Zhu, Y, Sun, F. | | Deposit date: | 2021-10-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
7VCI
 
 | | Structure of Xenopus laevis NPC nuclear ring asymmetric unit | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Tai, L, Zhu, Y, Sun, F. | | Deposit date: | 2021-09-03 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8.1 Å) | | Cite: | 8 angstrom structure of the outer rings of the Xenopus laevis nuclear pore complex obtained by cryo-EM and AI.
Protein Cell, 13, 2022
|
|
7WB4
 
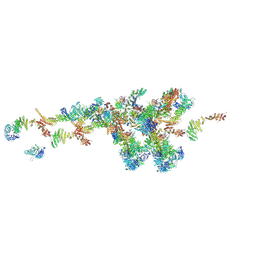 | | Cryo-EM structure of the NR subunit from X. laevis NPC | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Huang, G, Zhan, X, Shi, Y. | | Deposit date: | 2021-12-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Cryo-EM structure of the nuclear ring from Xenopus laevis nuclear pore complex.
Cell Res., 32, 2022
|
|
7WKK
 
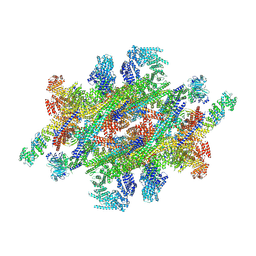 | | Cryo-EM structure of the IR subunit from X. laevis NPC | | Descriptor: | Aaas-prov protein, IL4I1 protein, MGC83295 protein, ... | | Authors: | Huang, G, Zhan, X, Shi, Y. | | Deposit date: | 2022-01-10 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the inner ring from the Xenopus laevis nuclear pore complex.
Cell Res., 32, 2022
|
|
7XP6
 
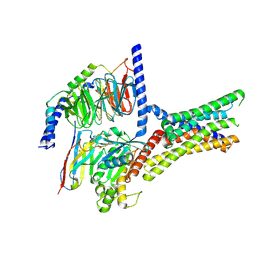 | | Cryo-EM structure of a class T GPCR in active state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP4
 
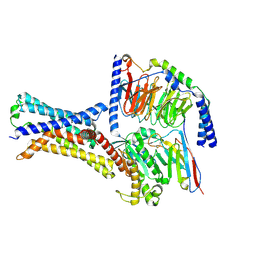 | | Cryo-EM structure of a class T GPCR in apo state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7XP5
 
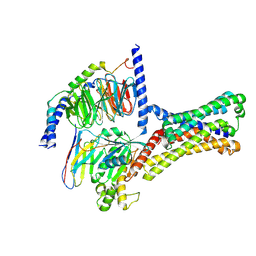 | | Cryo-EM structure of a class T GPCR in ligand-free state | | Descriptor: | Endoglucanase H,Taste receptor type 2 member 46,Bitter taste receptor T2R46, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Liu, Z.J, Hua, T, Xu, W.X, Wu, L.J. | | Deposit date: | 2022-05-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis for strychnine activation of human bitter taste receptor TAS2R46.
Science, 377, 2022
|
|
7UWF
 
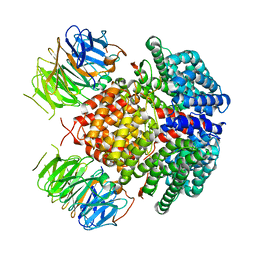 | | Human Rix1 sub-complex scaffold | | Descriptor: | Modulator of non-genomic activity of estrogen receptor, WD repeat-containing protein 18 | | Authors: | Gordon, J, Stanley, R.E. | | Deposit date: | 2022-05-03 | | Release date: | 2022-11-02 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM reveals the architecture of the PELP1-WDR18 molecular scaffold.
Nat Commun, 13, 2022
|
|
7UOH
 
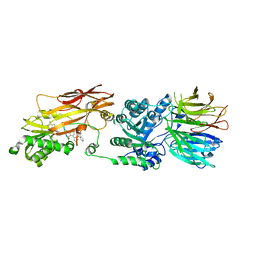 | | PRMT5/MEP50 crystal structure with MTA and an achiral, class 1, non-atropisomeric inhibitor bound | | Descriptor: | (2M)-2-[(4M)-4-{4-(aminomethyl)-1-oxo-8-[(2R)-oxolan-2-yl]-1,2-dihydrophthalazin-6-yl}-1-methyl-1H-pyrazol-5-yl]-1-benzothiophene-3-carbonitrile, 5'-DEOXY-5'-METHYLTHIOADENOSINE, Methylosome protein 50, ... | | Authors: | Gunn, R.J, Thomas, N.C, Lawson, J.D, Ivetac, A, Kulyk, S, Smith, C.R, Marx, M.A. | | Deposit date: | 2022-04-12 | | Release date: | 2022-08-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Design and evaluation of achiral, non-atropisomeric 4-(aminomethyl)phthalazin-1(2H)-one derivatives as novel PRMT5/MTA inhibitors.
Bioorg.Med.Chem., 71, 2022
|
|
