5JCP
 
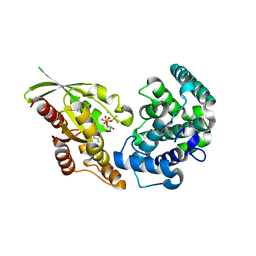 | | RhoGAP domain of ARAP3 in complex with RhoA in the transition state | | Descriptor: | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 3,Linker,Transforming protein RhoA, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bao, H, Li, F, Wang, C, Wang, N, Jiang, Y, Tang, Y, Wu, J, Shi, Y. | | Deposit date: | 2016-04-15 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis for the Specific Recognition of RhoA by the Dual GTPase-activating Protein ARAP3
J.Biol.Chem., 291, 2016
|
|
7RQY
 
 | | Cryo-EM structure of the full-length TRPV1 with RTx at 25 degrees Celsius, in an open state, class B | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
7RQX
 
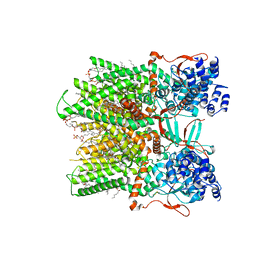 | | Cryo-EM structure of the full-length TRPV1 with RTx at 25 degrees Celsius, in an intermediate-open state, class A | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
7RQU
 
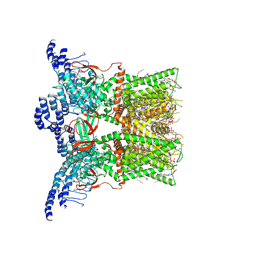 | | Cryo-EM structure of the full-length TRPV1 with RTx at 4 degrees Celsius, in a closed state, class I | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, SODIUM ION, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
7RQW
 
 | | Cryo-EM structure of the full-length TRPV1 with RTx at 4 degrees Celsius, in an open state, class III | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
7RQV
 
 | | Cryo-EM structure of the full-length TRPV1 with RTx at 4 degrees Celsius, in an intermediate-closed state, class II | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-08-08 | | Release date: | 2022-06-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Vanilloid-dependent TRPV1 opening trajectory from cryoEM ensemble analysis.
Nat Commun, 13, 2022
|
|
6JXX
 
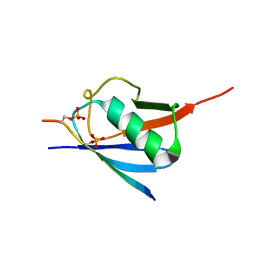 | | SUMO2 bound to phosphorylated SLS4-SIM peptide from ICP0 | | Descriptor: | Phosphorylated SLS4 from E3 ubiquitin ligase ICP0, Small ubiquitin-related modifier 2 | | Authors: | Hembram, D.S.S, Negi, H, Shet, D, Das, R. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Viral SUMO-Targeted Ubiquitin Ligase ICP0 is Phosphorylated and Activated by Host Kinase Chk2.
J.Mol.Biol., 432, 2020
|
|
6JXW
 
 | | Complex of SUMO2 bound SLS4 from ICP0. | | Descriptor: | SLS4-SIM from Ubiquitin E3 ligase ICP0, Small ubiquitin-related modifier 2 | | Authors: | Hembram, D.S.S, Negi, H, Shet, D, Das, R. | | Deposit date: | 2019-04-25 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Viral SUMO-Targeted Ubiquitin Ligase ICP0 is Phosphorylated and Activated by Host Kinase Chk2.
J.Mol.Biol., 432, 2020
|
|
7SI4
 
 | | CRYSTAL STRUCTURE OF EED WITH MRTX-2219 | | Descriptor: | (4S)-8-{4-[(dimethylamino)methyl]-2-methylphenyl}-5-{[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]amino}imidazo[1,2-c]pyrimidine-2-carbonitrile, FORMIC ACID, Polycomb protein EED | | Authors: | Gunn, R.J, Burns, A.C, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | CRYSTAL STRUCTURE OF EED WITH MRTX-2219
TO BE PUBLISHED
|
|
6KO6
 
 | | Crystal structure of AMPPNP bound Cka1 from C. neoformans | | Descriptor: | CMGC/CK2 protein kinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Cho, H.S, Yoo, Y. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of fungal pathogenicity-related casein kinase alpha subunit, Cka1, in the human fungal pathogen Cryptococcus neoformans.
Sci Rep, 9, 2019
|
|
7SI5
 
 | | CRYSTAL STRUCTURE OF EED WITH MRTX-1919 | | Descriptor: | (4R)-8-(1,3-dimethyl-1H-pyrazol-5-yl)-5-{[(5-fluoro-2,3-dihydro-1-benzofuran-4-yl)methyl]amino}imidazo[1,2-c]pyrimidine-2-carbonitrile, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Gunn, R.J, Burns, A.C, Lawson, J.D, Marx, M.A. | | Deposit date: | 2021-10-12 | | Release date: | 2022-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | CRYSTAL STRUCTURE OF EED WITH MRTX-1919
TO BE PUBLISHED
|
|
8COB
 
 | |
7TD3
 
 | | Sphingosine-1-phosphate receptor 1-Gi complex bound to S1P | | Descriptor: | (2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
7TD4
 
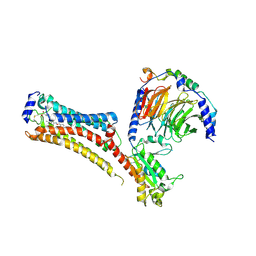 | | Sphingosine-1-phosphate receptor 1-Gi complex bound to Siponimod | | Descriptor: | 1-[[4-[(~{E})-~{N}-[[4-cyclohexyl-3-(trifluoromethyl)phenyl]methoxy]-~{C}-methyl-carbonimidoyl]-2-ethyl-phenyl]methyl]azetidine-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Liu, S, Paknejad, N, Zhu, L, Kihara, Y, Ray, D, Chun, J, Liu, W, Hite, R.K, Huang, X.Y. | | Deposit date: | 2021-12-30 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Differential activation mechanisms of lipid GPCRs by lysophosphatidic acid and sphingosine 1-phosphate.
Nat Commun, 13, 2022
|
|
6TDA
 
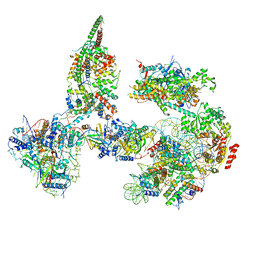 | | Structure of SWI/SNF chromatin remodeler RSC bound to a nucleosome | | Descriptor: | Actin-like protein ARP9, Actin-related protein 7, Chromatin structure-remodeling complex protein RSC58, ... | | Authors: | Wagner, F.R, Dienemann, C, Wang, H, Stuetzer, A, Tegunov, D, Urlaub, H, Cramer, P. | | Deposit date: | 2019-11-08 | | Release date: | 2020-03-18 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (15 Å) | | Cite: | Structure of SWI/SNF chromatin remodeller RSC bound to a nucleosome.
Nature, 579, 2020
|
|
8BJC
 
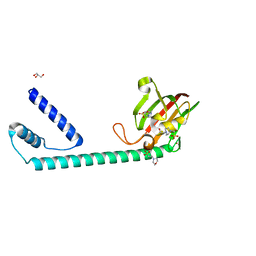 | | Full length structure of the apo-state LpMIP. | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Whittaker, J.J, Guskov, A, Goretzki, B, Hellmich, U.A. | | Deposit date: | 2022-11-03 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Legionella pneumophila macrophage infectivity potentiator protein appendage domains modulate protein dynamics and inhibitor binding.
Int.J.Biol.Macromol., 252, 2023
|
|
7S68
 
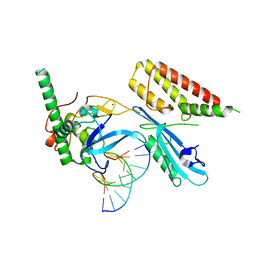 | | Structure of human PARP1 domains (Zn1, Zn3, WGR and HD) bound to a DNA double strand break. | | Descriptor: | DNA (5'-D(*GP*CP*CP*TP*GP*CP*AP*GP*GP*C)-3'), Fusion of PARP1 zinc fingers 1 and 3 (Zn1, Zn3), ... | | Authors: | Rouleau-Turcotte, E, Pascal, J.M. | | Deposit date: | 2021-09-13 | | Release date: | 2022-06-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Captured snapshots of PARP1 in the active state reveal the mechanics of PARP1 allostery.
Mol.Cell, 82, 2022
|
|
7KW7
 
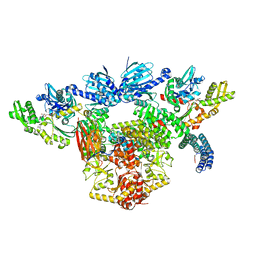 | | Atomic cryoEM structure of Hsp90-Hsp70-Hop-GR | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Glucocorticoid receptor, Heat shock 70 kDa protein 1A, ... | | Authors: | Wang, R.Y, Noddings, C.M, Kirschke, E, Myasnikov, A, Johnson, J.L, Agard, D.A. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structure of Hsp90-Hsp70-Hop-GR reveals the Hsp90 client-loading mechanism.
Nature, 601, 2022
|
|
8CDJ
 
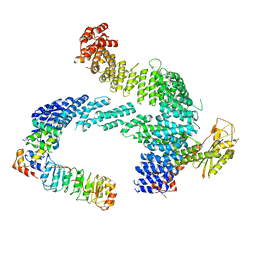 | | CAND1 b-hairpin++-SCF-SKP2 CAND1 rolling SCF engaged | | Descriptor: | Cullin-1, Cullin-associated NEDD8-dissociated protein 1, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Baek, K, Schulman, B.A. | | Deposit date: | 2023-01-31 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Systemwide disassembly and assembly of SCF ubiquitin ligase complexes.
Cell, 186, 2023
|
|
8CDK
 
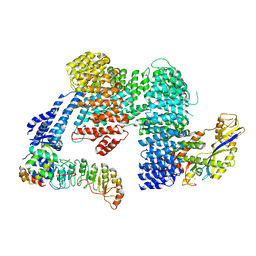 | |
7Y8R
 
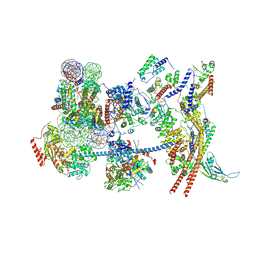 | | The nucleosome-bound human PBAF complex | | Descriptor: | ACTB protein (Fragment), ADENOSINE-5'-DIPHOSPHATE, AT-rich interactive domain-containing protein 2, ... | | Authors: | Wang, L, Yu, J, Yu, Z, Wang, Q, He, S, Xu, Y. | | Deposit date: | 2022-06-24 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of nucleosome-bound human PBAF complex.
Nat Commun, 13, 2022
|
|
9IPU
 
 | |
8CXI
 
 | | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy | | Descriptor: | A11 heavy chain, A11 light chain, Ankyrin repeat family A protein 2,Envelope E protein, ... | | Authors: | Liu, W, Zhang, X.K, Gong, D.Y, Dai, X.H, Sharma, A, Zhang, T.H, Rey, F, Zhou, Z.H. | | Deposit date: | 2022-05-21 | | Release date: | 2023-06-07 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of Zika Virus in Complex with Antibodies Targeting E Dimer Epitopes and Basis for Neutralization Efficacy
To Be Published
|
|
6GEN
 
 | | Chromatin remodeller-nucleosome complex at 4.5 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Willhoft, O, Chua, E.Y.D, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2018-04-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of the yeast SWR1-nucleosome complex.
Science, 362, 2018
|
|
6GEJ
 
 | | Chromatin remodeller-nucleosome complex at 3.6 A resolution. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Willhoft, O, Chua, E.Y.D, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2018-04-26 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of the yeast SWR1-nucleosome complex.
Science, 362, 2018
|
|
