19GS
 
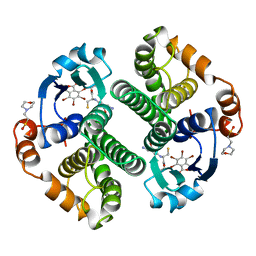 | | Glutathione s-transferase p1-1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3,3'-(4,5,6,7-TETRABROMO-3-OXO-1(3H)-ISOBENZOFURANYLIDENE)BIS [6-HYDROXYBENZENESULFONIC ACID]ANION, GLUTATHIONE, ... | | Authors: | Oakley, A.J, Lo Bello, M, Parker, M.W. | | Deposit date: | 1997-12-14 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
102M
 
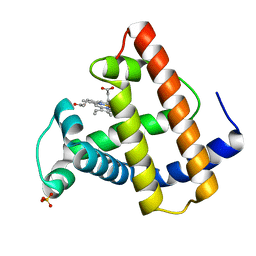 | | SPERM WHALE MYOGLOBIN H64A AQUOMET AT PH 9.0 | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-15 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
1A1S
 
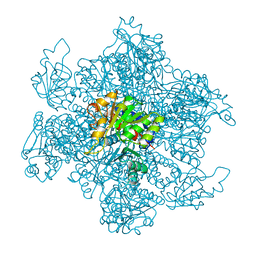 | | ORNITHINE CARBAMOYLTRANSFERASE FROM PYROCOCCUS FURIOSUS | | Descriptor: | ORNITHINE CARBAMOYLTRANSFERASE | | Authors: | Villeret, V, Clantin, B, Tricot, C, Legrain, C, Roovers, M, Stalon, V, Glansdorff, N, Van Beeumen, J. | | Deposit date: | 1997-12-15 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of Pyrococcus furiosus ornithine carbamoyltransferase reveals a key role for oligomerization in enzyme stability at extremely high temperatures.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1A1R
 
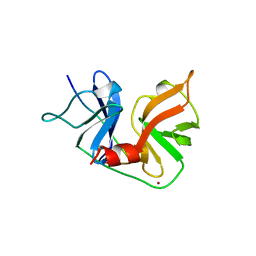 | | HCV NS3 PROTEASE DOMAIN:NS4A PEPTIDE COMPLEX | | Descriptor: | NS3 PROTEIN, NS4A PROTEIN, ZINC ION | | Authors: | Kim, J.L, Morgenstern, K.A, Lin, C, Fox, T, Dwyer, M.D, Landro, J.A, Chambers, S.P, Markland, W, Lepre, C.A, O'Malley, E.T, Harbeson, S.L, Rice, C.M, Murcko, M.A, Caron, P.R, Thomson, J.A. | | Deposit date: | 1997-12-15 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide.
Cell(Cambridge,Mass.), 87, 1996
|
|
1A1T
 
 | | STRUCTURE OF THE HIV-1 NUCLEOCAPSID PROTEIN BOUND TO THE SL3 PSI-RNA RECOGNITION ELEMENT, NMR, 25 STRUCTURES | | Descriptor: | NUCLEOCAPSID PROTEIN, SL3 STEM-LOOP RNA, ZINC ION | | Authors: | De Guzman, R.N, Wu, Z.R, Stalling, C.C, Pappalardo, L, Borer, P.N, Summers, M.F. | | Deposit date: | 1997-12-15 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element.
Science, 279, 1998
|
|
2DEF
 
 | |
103M
 
 | | SPERM WHALE MYOGLOBIN H64A N-BUTYL ISOCYANIDE AT PH 9.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-16 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
1A1U
 
 | | SOLUTION STRUCTURE DETERMINATION OF A P53 MUTANT DIMERIZATION DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | P53 | | Authors: | Mccoy, M.A, Stavridi, E.S, Waterman, J.L.F, Wieczorek, A, Opella, S.J, Halezonetis, T.D. | | Deposit date: | 1997-12-16 | | Release date: | 1998-04-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Hydrophobic side-chain size is a determinant of the three-dimensional structure of the p53 oligomerization domain.
EMBO J., 16, 1997
|
|
20GS
 
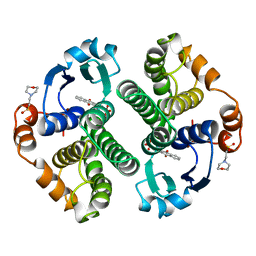 | | GLUTATHIONE S-TRANSFERASE P1-1 COMPLEXED WITH CIBACRON BLUE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CIBACRON BLUE, GLUTATHIONE S-TRANSFERASE | | Authors: | Oakley, A.J, Lo Bello, M, Nuccetelli, M, Mazzetti, A.P, Parker, M.W. | | Deposit date: | 1997-12-16 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The ligandin (non-substrate) binding site of human Pi class glutathione transferase is located in the electrophile binding site (H-site).
J.Mol.Biol., 291, 1999
|
|
1VGH
 
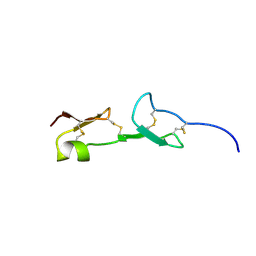 | | HEPARIN-BINDING DOMAIN FROM VASCULAR ENDOTHELIAL GROWTH FACTOR, NMR, 20 STRUCTURES | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR-165 | | Authors: | Fairbrother, W.J, Champe, M.A, Christinger, H.W, Keyt, B.A, Starovasnik, M.A. | | Deposit date: | 1997-12-17 | | Release date: | 1998-04-08 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the heparin-binding domain of vascular endothelial growth factor.
Structure, 6, 1998
|
|
365D
 
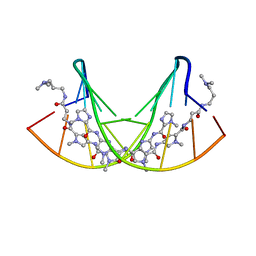 | | STRUCTURAL BASIS FOR G C RECOGNITION IN THE DNA MINOR GROOVE | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*GP*(CBR)P*CP*TP*GP*G)-3'), IMIDAZOLE-PYRROLE POLYAMIDE | | Authors: | Kielkopf, C.L, Baird, E.E, Dervan, P.B, Rees, D.C. | | Deposit date: | 1997-12-17 | | Release date: | 1998-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for G.C recognition in the DNA minor groove.
Nat.Struct.Biol., 5, 1998
|
|
1A1V
 
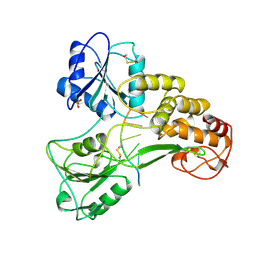 | | HEPATITIS C VIRUS NS3 HELICASE DOMAIN COMPLEXED WITH SINGLE STRANDED SDNA | | Descriptor: | DNA (5'-D(*UP*UP*UP*UP*UP*UP*UP*U)-3'), PROTEIN (NS3 PROTEIN), SULFATE ION | | Authors: | Kim, J.L, Morgenstern, K.A, Griffith, J.P, Dwyer, M.D, Thomson, J.A, Murcko, M.A, Lin, C, Caron, P.R. | | Deposit date: | 1997-12-17 | | Release date: | 1999-01-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Hepatitis C virus NS3 RNA helicase domain with a bound oligonucleotide: the crystal structure provides insights into the mode of unwinding.
Structure, 6, 1998
|
|
2VGH
 
 | | HEPARIN-BINDING DOMAIN FROM VASCULAR ENDOTHELIAL GROWTH FACTOR, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | VASCULAR ENDOTHELIAL GROWTH FACTOR-165 | | Authors: | Fairbrother, W.J, Champe, M.A, Christinger, H.W, Keyt, B.A, Starovasnik, M.A. | | Deposit date: | 1997-12-17 | | Release date: | 1998-04-08 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the heparin-binding domain of vascular endothelial growth factor.
Structure, 6, 1998
|
|
105M
 
 | | SPERM WHALE MYOGLOBIN N-BUTYL ISOCYANIDE AT PH 9.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-18 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
104M
 
 | | SPERM WHALE MYOGLOBIN N-BUTYL ISOCYANIDE AT PH 7.0 | | Descriptor: | MYOGLOBIN, N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Smith, R.D, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1997-12-18 | | Release date: | 1998-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Correlations between Bound N-Alkyl Isocyanide Orientations and Pathways for Ligand Binding in Recombinant Myoglobins
Thesis, Rice, 1999
|
|
1A1W
 
 | | FADD DEATH EFFECTOR DOMAIN, F25Y MUTANT, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FADD PROTEIN | | Authors: | Eberstadt, M, Huang, B, Chen, Z, Meadows, R.P, Ng, C, Fesik, S.W. | | Deposit date: | 1997-12-18 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and mutagenesis of the FADD (Mort1) death-effector domain.
Nature, 392, 1998
|
|
2HGF
 
 | | HAIRPIN LOOP CONTAINING DOMAIN OF HEPATOCYTE GROWTH FACTOR, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HEPATOCYTE GROWTH FACTOR | | Authors: | Zhou, H, Mazzulla, M.J, Kaufman, J.D, Stahl, S.J, Wingfield, P.T, Rubin, J.S, Bottaro, D.P, Byrd, R.A. | | Deposit date: | 1997-12-18 | | Release date: | 1998-06-24 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the N-terminal domain of hepatocyte growth factor reveals a potential heparin-binding site.
Structure, 6, 1998
|
|
1ZDJ
 
 | | STRUCTURE OF BACTERIOPHAGE COAT PROTEIN-LOOP RNA COMPLEX | | Descriptor: | PROTEIN (MS2 PROTEIN CAPSID), RNA (5'-R(*GP*GP*AP*UP*CP*AP*CP*C)-3') | | Authors: | Grahn, E, Stonehouse, N, Valegard, K, Vandenworm, S, Liljas, L. | | Deposit date: | 1997-12-18 | | Release date: | 1998-07-08 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic studies of RNA hairpins in complexes with recombinant MS2 capsids: implications for binding requirements.
RNA, 5, 1999
|
|
1A1Z
 
 | | FADD DEATH EFFECTOR DOMAIN, F25G MUTANT, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | FADD PROTEIN | | Authors: | Eberstadt, M, Huang, B, Chen, Z, Meadows, R.P, Ng, C, Fesik, S.W. | | Deposit date: | 1997-12-18 | | Release date: | 1998-12-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure and mutagenesis of the FADD (Mort1) death-effector domain.
Nature, 392, 1998
|
|
1OAK
 
 | |
1A1X
 
 | | CRYSTAL STRUCTURE OF MTCP-1 INVOLVED IN T CELL MALIGNANCIES | | Descriptor: | HMTCP-1 | | Authors: | Fu, Z.Q, Dubois, G.C, Song, S.P, Kulikovskaya, I, Virgilio, L, Rothstein, J, Croce, C.M, Weber, I.T, Harrison, R.W. | | Deposit date: | 1997-12-18 | | Release date: | 1998-05-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of MTCP-1: implications for role of TCL-1 and MTCP-1 in T cell malignancies.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1A12
 
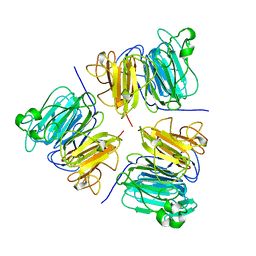 | | REGULATOR OF CHROMOSOME CONDENSATION (RCC1) OF HUMAN | | Descriptor: | REGULATOR OF CHROMOSOME CONDENSATION 1 | | Authors: | Renault, L, Nassar, N, Vetter, I, Becker, J, Roth, M, Wittinghofer, A. | | Deposit date: | 1997-12-19 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller.
Nature, 392, 1998
|
|
368D
 
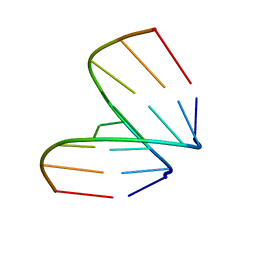 | | STRUCTURAL VARIABILITY OF A-DNA IN CRYSTALS OF THE OCTAMER D(PCPCPCPGPCPGPGPG) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*GP*CP*GP*GP*G)-3') | | Authors: | Fernandez, L.G, Subirana, J.A, Verdaguer, N, Pyshnyi, D, Campos, L, Malinina, L. | | Deposit date: | 1997-12-19 | | Release date: | 1998-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural variability of A-DNA in crystals of the octamer d(pCpCpCpGpCpGpGpG)
J.Biomol.Struct.Dyn., 15, 1997
|
|
367D
 
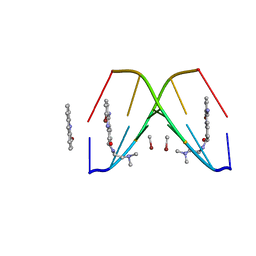 | | 1.2 A STRUCTURE DETERMINATION OF THE D(CG(5-BRU)ACG)2/5-BROMO-9-AMINO-DACA COMPLEX | | Descriptor: | 5'-D(*CP*GP*(BRU)P*AP*CP*G)-3', 5-BROMO-9-AMINO-N-ETHYL(DIAMINOMETHYL)ACRIDINE-4-CARBOXAMIDE, BROMIDE ION | | Authors: | Todd, A.K, Adams, A, Thorpe, J.H, Denny, W.A, Cardin, C.J. | | Deposit date: | 1997-12-19 | | Release date: | 2003-03-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Major groove binding and 'DNA-induced' fit in the intercalation of a derivative of the mixed topoisomerase I/II poison N-(2-(dimethlyamino)ethyl)acridine-4-carboxamide (DACA) into DNA: X-ray structure complexed to d(CG(5Br-U)ACG)2 at 1.3-angstrom resolution
J.Med.Chem., 42, 1999
|
|
366D
 
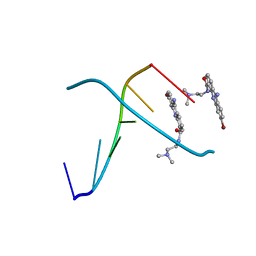 | | 1.3 A STRUCTURE DETERMINATION OF THE D(CG(5-BRU)ACG)2/6-BROMO-9-AMINO-DACA COMPLEX | | Descriptor: | 6-BROMO-9-AMINO-N-ETHYL(DIAMINOMETHYL)ACRIDINE-4-CARBOXAMIDE, DNA (5'-D(*CP*GP*(BRU)P*AP*CP*G)-3') | | Authors: | Todd, A.K, Adams, A, Thorpe, J.H, Denny, W.A, Cardin, C.J. | | Deposit date: | 1997-12-19 | | Release date: | 1999-04-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Major groove binding and 'DNA-induced' fit in the intercalation of a derivative of the mixed topoisomerase I/II poison N-(2-(dimethylamino)ethyl)acridine-4-carboxamide (DACA) into DNA: X-ray structure complexed to d(CG(5-BrU)ACG)2 at 1.3-A resolution.
J.Med.Chem., 42, 1999
|
|
