7ACF
 
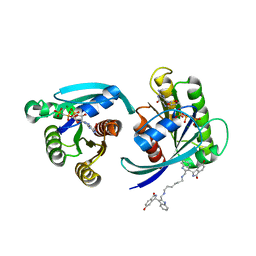 | | CRYSTAL STRUCTURE OF CRYSTAL FORM 2 OF AN ACTIVE KRAS G12D (GPPCP) DIMER IN COMPLEX WITH BI-5747 | | Descriptor: | (3~{S})-5-oxidanyl-3-[2-[[6-[[3-[(1~{S})-6-oxidanyl-3-oxidanylidene-1,2-dihydroisoindol-1-yl]-1~{H}-indol-2-yl]methylamino]hexylamino]methyl]-1~{H}-indol-3-yl]-2,3-dihydroisoindol-1-one, GTPase KRas, MAGNESIUM ION, ... | | Authors: | Kessler, D. | | Deposit date: | 2020-09-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Reply to Tran et al.: Dimeric KRAS protein-protein interaction stabilizers.
Proc. Natl. Acad. Sci. U.S.A., 117, 2020
|
|
1ACF
 
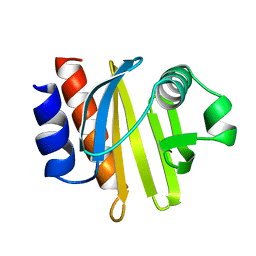 | | ACANTHAMOEBA CASTELLANII PROFILIN IB | | Descriptor: | PROFILIN I | | Authors: | Fedorov, A.A, Magnus, K.A, Graupe, M.H, Lattman, E.E, Pollard, T.D, Almo, S.C. | | Deposit date: | 1994-07-29 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structures of isoforms of the actin-binding protein profilin that differ in their affinity for phosphatidylinositol phosphates.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
8ACF
 
 | |
2ACF
 
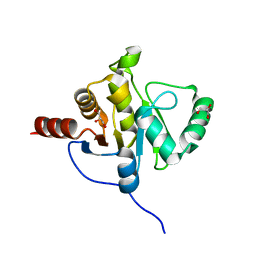 | | NMR STRUCTURE OF SARS-COV NON-STRUCTURAL PROTEIN NSP3A (SARS1) FROM SARS CORONAVIRUS | | Descriptor: | GLYCEROL, Replicase polyprotein 1ab | | Authors: | Saikatendu, K.S, Joseph, J.S, Subramanian, V, Neuman, B.W, Buchmeier, M.J, Stevens, R.C, Kuhn, P, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2005-07-18 | | Release date: | 2006-02-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1''-phosphate dephosphorylation by a conserved domain of nsP3.
Structure, 13, 2005
|
|
3ACF
 
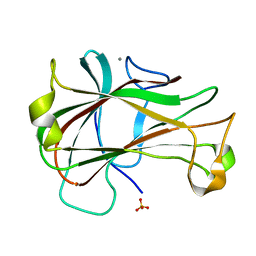 | | Crystal Structure of Carbohydrate-Binding Module Family 28 from Clostridium josui Cel5A in a ligand-free form | | Descriptor: | Beta-1,4-endoglucanase, CALCIUM ION, SULFATE ION | | Authors: | Tsukimoto, K, Takada, R, Araki, Y, Suzuki, K, Karita, S, Wakagi, T, Shoun, H, Watanabe, T, Fushinobu, S. | | Deposit date: | 2010-01-04 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of cellooligosaccharides by a family 28 carbohydrate-binding module.
Febs Lett., 584, 2010
|
|
7TY1
 
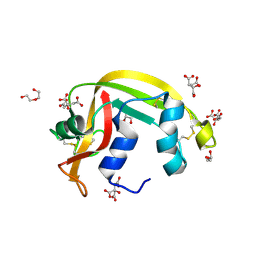 | | Crystal structure of apo eosinophil cationic protein (ribonuclease 3) from Macaca fascicularis (MfECP) | | Descriptor: | CITRIC ACID, DI(HYDROXYETHYL)ETHER, Eosinophil cationic protein, ... | | Authors: | Tran, T.T.Q, Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2022-02-11 | | Release date: | 2023-08-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ancestral sequence reconstruction dissects structural and functional differences among eosinophil ribonucleases.
J.Biol.Chem., 300, 2024
|
|
5ACG
 
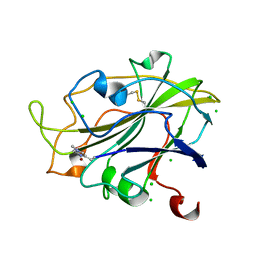 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
5ACI
 
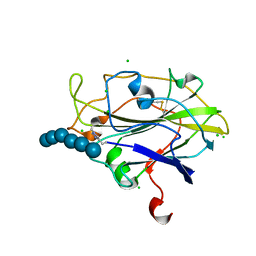 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johanson, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
5ACJ
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
5ACH
 
 | | X-ray Structure of LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2015-08-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The molecular basis of polysaccharide cleavage by lytic polysaccharide monooxygenases.
Nat. Chem. Biol., 12, 2016
|
|
8AHE
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | SULFATE ION, UDP-N-acetylglucosamine 2-epimerase, ~{N},5-dimethyl-1-phenyl-pyrazole-4-sulfonamide | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.108 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
8AHF
 
 | | PAC-FragmentDEL: Photoactivated covalent capture of DNA encoded fragments for hit discovery | | Descriptor: | (2~{R},4~{S})-4-[bis(fluoranyl)methoxy]-~{N}-methyl-1-(2~{H}-pyrazolo[4,3-b]pyridin-6-ylcarbonyl)pyrrolidine-2-carboxamide, SULFATE ION, UDP-N-acetylglucosamine 2-epimerase | | Authors: | Baker, L.M, Murray, J.B, Hubbard, R.E. | | Deposit date: | 2022-07-21 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.271 Å) | | Cite: | PAC-FragmentDEL - photoactivated covalent capture of DNA-encoded fragments for hit discovery.
Rsc Med Chem, 13, 2022
|
|
1AYX
 
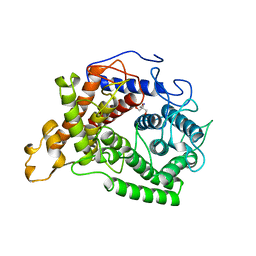 | | CRYSTAL STRUCTURE OF GLUCOAMYLASE FROM SACCHAROMYCOPSIS FIBULIGERA AT 1.7 ANGSTROMS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUCOAMYLASE | | Authors: | Sevcik, J, Hostinova, E, Gasperik, J, Solovicova, A, Wilson, K.S, Dauter, Z. | | Deposit date: | 1997-11-12 | | Release date: | 1998-05-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of glucoamylase from Saccharomycopsis fibuligera at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1UCY
 
 | |
1A8T
 
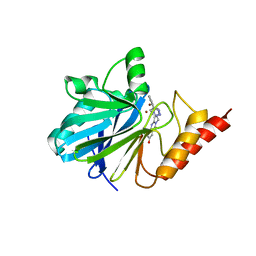 | | METALLO-BETA-LACTAMASE IN COMPLEX WITH L-159,061 | | Descriptor: | 2-BUTYL-6-HYDROXY-3-[2'-(1H-TETRAZOL-5-YL)-BIPHENYL-4-YLMETHYL]-3H-QUINAZOLIN-4-ONE, METALLO-BETA-LACTAMASE, ZINC ION | | Authors: | Fitzgerald, P.M.D, Toney, J.H, Grover, N, Vanderwall, D. | | Deposit date: | 1998-03-23 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Antibiotic sensitization using biphenyl tetrazoles as potent inhibitors of Bacteroides fragilis metallo-beta-lactamase.
Chem.Biol., 5, 1998
|
|
1A7T
 
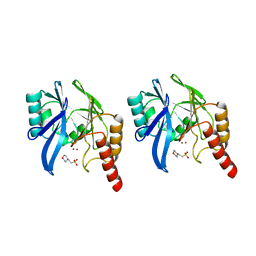 | | METALLO-BETA-LACTAMASE WITH MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, METALLO-BETA-LACTAMASE, SODIUM ION, ... | | Authors: | Fitzgerald, P.M.D, Wu, J.K, Toney, J.H. | | Deposit date: | 1998-03-17 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Unanticipated inhibition of the metallo-beta-lactamase from Bacteroides fragilis by 4-morpholineethanesulfonic acid (MES): a crystallographic study at 1.85-A resolution.
Biochemistry, 37, 1998
|
|
8H3X
 
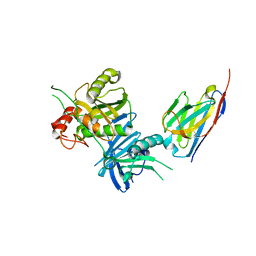 | | Bacteroide Fragilis Toxin in complex with nanobody 282 | | Descriptor: | Fragilysin, ZINC ION, nanobody 282 | | Authors: | Wen, Y, Guo, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Screening and epitope characterization of diagnostic nanobody against total and activated Bacteroides fragilis toxin.
Front Immunol, 14, 2023
|
|
8H3Y
 
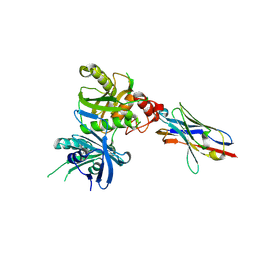 | | Bacteroide Fragilis Toxin in complex with nanobody 327 | | Descriptor: | Fragilysin, Nanobody 327, ZINC ION | | Authors: | Wen, Y, Guo, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-02-08 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Screening and epitope characterization of diagnostic nanobody against total and activated Bacteroides fragilis toxin.
Front Immunol, 14, 2023
|
|
1KR3
 
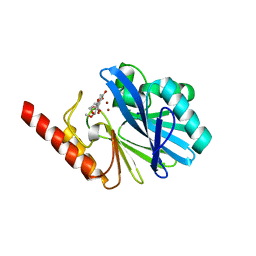 | | Crystal Structure of the Metallo beta-Lactamase from Bacteroides fragilis (CfiA) in Complex with the Tricyclic Inhibitor SB-236050. | | Descriptor: | 7,8-DIHYDROXY-1-METHOXY-3-METHYL-10-OXO-4,10-DIHYDRO-1H,3H-PYRANO[4,3-B]CHROMENE-9-CARBOXYLIC ACID, SODIUM ION, ZINC ION, ... | | Authors: | Payne, D.J, Hueso-Rodrguez, J.A, Boyd, H, Concha, N.O, Janson, C.A, Gilpin, M, Bateson, J.H, Cheever, C, Niconovich, N.L, Pearson, S, Rittenhouse, S, Tew, D, Dez, E, Prez, P, de la Fuente, J, Rees, M, Rivera-Sagredo, A. | | Deposit date: | 2002-01-08 | | Release date: | 2003-01-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a series of tricyclic natural products as potent broad-spectrum inhibitors of metallo-beta-lactamases
ANTIMICROB.AGENTS CHEMOTHER., 46, 2002
|
|
1HLK
 
 | | METALLO-BETA-LACTAMASE FROM BACTEROIDES FRAGILIS IN COMPLEX WITH A TRICYCLIC INHIBITOR | | Descriptor: | 7,8-DIHYDROXY-1-METHOXY-3-METHYL-10-OXO-4,10-DIHYDRO-1H,3H-PYRANO[4,3-B]CHROMENE-9-CARBOXYLIC ACID, BETA-LACTAMASE, TYPE II, ... | | Authors: | Payne, D.J, Hueso-Rodriguez, J.A, Boyd, H, Concha, N.O, Janson, C.A, Gilpin, M, Bateson, J.H, Chever, C, Niconovich, N.L, Pearson, S, Rittenhouse, S, Tew, D, Diez, E, Perez, P, de la Fuente, J, Rees, M, Rivera-Sagredo, A. | | Deposit date: | 2000-12-01 | | Release date: | 2001-11-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Identification of a series of tricyclic natural products as potent broad spectrum inhibitors of metallo-beta-lactamases
Antimicrob.Agents Chemother., 46, 2002
|
|
4ZNB
 
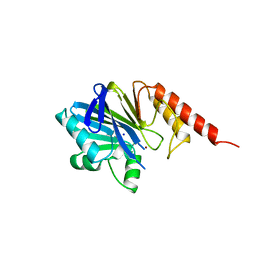 | | METALLO-BETA-LACTAMASE (C181S MUTANT) | | Descriptor: | METALLO-BETA-LACTAMASE, SODIUM ION, ZINC ION | | Authors: | Li, Z, Herzberg, O. | | Deposit date: | 1998-10-20 | | Release date: | 1999-06-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural consequences of the active site substitution Cys181 --> Ser in metallo-beta-lactamase from Bacteroides fragilis.
Protein Sci., 8, 1999
|
|
1ZNB
 
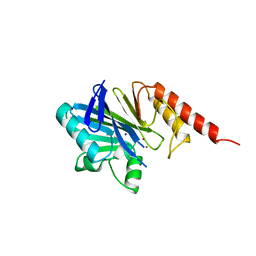 | | METALLO-BETA-LACTAMASE | | Descriptor: | METALLO-BETA-LACTAMASE, SODIUM ION, ZINC ION | | Authors: | Concha, N.O, Herzberg, O. | | Deposit date: | 1996-06-06 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the wide-spectrum binuclear zinc beta-lactamase from Bacteroides fragilis.
Structure, 4, 1996
|
|
4O6Y
 
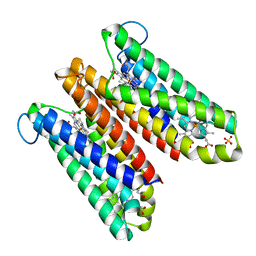 | | Crystal Structure of Cytochrome b561 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Probable transmembrane ascorbate ferrireductase 2, SULFATE ION | | Authors: | Lu, P, Ma, D, Yan, C, Gong, X, Du, M, Shi, Y. | | Deposit date: | 2013-12-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4O79
 
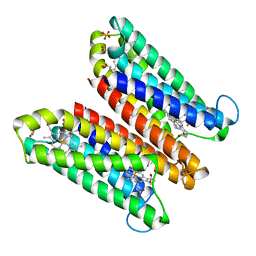 | | Crystal Structure of Ascorbate-bound Cytochrome b561, crystal soaked in 1 M L-ascorbate for 10 minutes | | Descriptor: | ASCORBIC ACID, PROTOPORPHYRIN IX CONTAINING FE, Probable transmembrane ascorbate ferrireductase 2, ... | | Authors: | Lu, P, Ma, D, Yan, C, Gong, X, Du, M, Shi, Y. | | Deposit date: | 2013-12-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4O7G
 
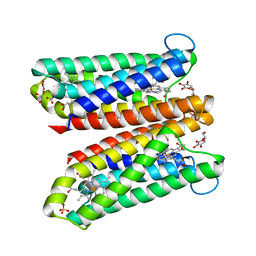 | | Crystal Structure of Ascorbate-bound Cytochrome b561, crystal soaked in 1 M L-ascorbate for 40 minutes | | Descriptor: | ASCORBIC ACID, PROTOPORPHYRIN IX CONTAINING FE, Probable transmembrane ascorbate ferrireductase 2, ... | | Authors: | Lu, P, Ma, D, Yan, C, Gong, X, Du, M, Shi, Y. | | Deposit date: | 2013-12-24 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.211 Å) | | Cite: | Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
