7B25
 
 | |
5FR7
 
 | | Erwinia amylovora AmyR amylovoran repressor, a member of the YbjN protein family | | Descriptor: | AMYR | | Authors: | Bartho, J.D, Bellini, D, Wuerges, J, Demitri, N, Walsh, M, Benini, S. | | Deposit date: | 2015-12-16 | | Release date: | 2017-01-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The crystal structure of Erwinia amylovora AmyR, a member of the YbjN protein family, shows similarity to type III secretion chaperones but suggests different cellular functions.
PLoS ONE, 12, 2017
|
|
2V57
 
 | | Crystal structure of the TetR-like transcriptional regulator LfrR from Mycobacterium smegmatis in complex with proflavine | | Descriptor: | ISOPROPYL ALCOHOL, PROFLAVIN, SULFATE ION, ... | | Authors: | Bellinzoni, M, Buroni, S, Riccardi, G, De Rossi, E, Alzari, P.M. | | Deposit date: | 2008-10-02 | | Release date: | 2009-10-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Plasticity and Distinct Drug-Binding Modes of Lfrr, a Mycobacterial Efflux Pump Regulator.
J.Bacteriol., 191, 2009
|
|
1VPW
 
 | | STRUCTURE OF THE PURR MUTANT, L54M, BOUND TO HYPOXANTHINE AND PURF OPERATOR DNA | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), HYPOXANTHINE, PURINE REPRESSOR | | Authors: | Arvidson, D.N, Lu, F, Faber, C, Zalkin, H, Brennan, R.G. | | Deposit date: | 1998-02-27 | | Release date: | 1998-06-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of PurR mutant L54M shows an alternative route to DNA kinking.
Nat.Struct.Biol., 5, 1998
|
|
6O8L
 
 | |
6O8K
 
 | | Crystal Structure of apo and reduced Sulfide-responsive transcriptional repressor (SqrR) from Rhodobacter capsulatus. | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator, ... | | Authors: | Capdevila, D.A, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2019-03-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural basis for persulfide-sensing specificity in a transcriptional regulator.
Nat.Chem.Biol., 17, 2021
|
|
6O8N
 
 | |
6O8M
 
 | | Crystal Structure of C9S apo Sulfide-responsive transcriptional repressor (SqrR) from Rhodobacter capsulated bound to diamide (tetramethylazodicarboxamide). | | Descriptor: | N~1~,N~1~,N~2~,N~2~-tetramethylhydrazine-1,2-dicarboxamide, Transcriptional regulator, ArsR family | | Authors: | Capdevila, D.A, Gonzalez-Gutierrez, G, Giedroc, D.P. | | Deposit date: | 2019-03-11 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural basis for persulfide-sensing specificity in a transcriptional regulator.
Nat.Chem.Biol., 17, 2021
|
|
6T46
 
 | | Structure of the Rap conjugation gene regulator of the plasmid pLS20 in complex with the Phr* peptide | | Descriptor: | CHLORIDE ION, Quorum-sensing secretion protein (processed), Response regulator aspartate phosphatase, ... | | Authors: | Crespo, I, Bernardo, N, Meijer, W.J.J, Boer, D.R. | | Deposit date: | 2019-10-12 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Inactivation of the dimeric RappLS20 anti-repressor of the conjugation operon is mediated by peptide-induced tetramerization.
Nucleic Acids Res., 48, 2020
|
|
6T3H
 
 | |
2UXN
 
 | | Structural Basis of Histone Demethylation by LSD1 Revealed by Suicide Inactivation | | Descriptor: | CHLORIDE ION, DIHYDROFLAVINE-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Yang, M, Culhane, J.C, Szewczuk, L.M, Gocke, C.B, Brautigam, C.A, Tomchick, D.R, Machius, M, Cole, P.A, Yu, H. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural Basis of Histone Demethylation by Lsd1 Revealed by Suicide Inactivation.
Nat.Struct.Mol.Biol., 14, 2007
|
|
5MWO
 
 | | Structure of Mycobacterium Tuberculosis Transcriptional Regulatory Repressor Protein (EthR) in complex with fragment 7E8. | | Descriptor: | (5-methyl-1-benzothiophen-2-yl)methanol, 1,2-ETHANEDIOL, HTH-type transcriptional regulator EthR | | Authors: | Mendes, V, Chan, D.S.-H, Thomas, S.E, McConnell, B, Matak-Vinkovic, D, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2017-01-18 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Fragment Screening against the EthR-DNA Interaction by Native Mass Spectrometry.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5ICJ
 
 | | Crystal structure of the Mycobacterium tuberculosis transcriptional repressor EthR2 in complex with BDM41420 | | Descriptor: | 4,4,4-trifluoro-1-(3-phenyl-1-oxa-2,8-diazaspiro[4.5]dec-2-en-8-yl)butan-1-one, Probable transcriptional regulatory protein | | Authors: | Wohlkonig, A, Remaut, H, Tanina, A, Meyer, F, Willand, N, Baulard, A.R, Wintjens, R. | | Deposit date: | 2016-02-23 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Reversion of antibiotic resistance in Mycobacterium tuberculosis by spiroisoxazoline SMARt-420.
Science, 355, 2017
|
|
5MXK
 
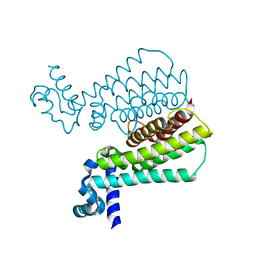 | | Structure of Mycobacterium Tuberculosis Transcriptional Regulatory Repressor Protein (EthR) in complex with fragment 7G9. | | Descriptor: | 1,2-ETHANEDIOL, HTH-type transcriptional regulator EthR, ~{N}-(5-oxidanylidene-7,8-dihydro-6~{H}-naphthalen-2-yl)ethanamide | | Authors: | Mendes, V, Chan, D.S.-H, Thomas, S.E, McConnell, B, Matak-Vinkovic, D, Coyne, A.G, Abell, C, Blundell, T.L. | | Deposit date: | 2017-01-23 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Fragment Screening against the EthR-DNA Interaction by Native Mass Spectrometry.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5N08
 
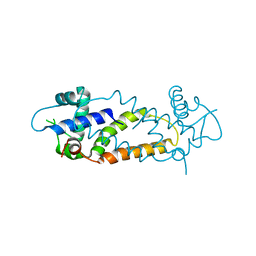 | |
4ZA6
 
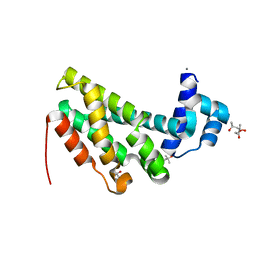 | |
6QDW
 
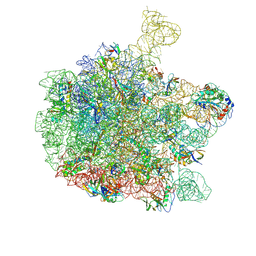 | | Cryo-EM structure of the 50S ribosomal subunit at 2.83 Angstroms with modeled GBC SecM peptide | | Descriptor: | 23S rRNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Schulte, L, Reitz, J, Hodirnau, V.V, Kudlinzki, D, Mao, J, Glaubitz, C, Frangakis, A, Schwalbe, H. | | Deposit date: | 2019-01-03 | | Release date: | 2020-01-15 | | Last modified: | 2020-12-02 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cysteine oxidation and disulfide formation in the ribosomal exit tunnel.
Nat Commun, 11, 2020
|
|
2HGV
 
 | |
2GX5
 
 | | N-terminal GAF domain of transcriptional pleiotropic repressor CodY | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, GLYCEROL, GTP-sensing transcriptional pleiotropic repressor codY, ... | | Authors: | Wilkinson, A.J, Levdikov, V.M, Blagova, E.V. | | Deposit date: | 2006-05-08 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in gram-positive bacteria.
J.Biol.Chem., 281, 2006
|
|
5YEG
 
 | | Crystal structure of CTCF ZFs4-8-Hs5-1a complex | | Descriptor: | DNA (5'-D(*AP*CP*TP*TP*TP*AP*AP*CP*CP*AP*GP*CP*AP*GP*AP*GP*GP*GP*CP*G)-3'), DNA (5'-D(*TP*CP*GP*CP*CP*CP*TP*CP*TP*GP*CP*TP*GP*GP*TP*TP*AP*AP*AP*G)-3'), Transcriptional repressor CTCF, ... | | Authors: | Yin, M, Wang, J, Wang, M, Li, X. | | Deposit date: | 2017-09-17 | | Release date: | 2017-11-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites
Cell Res., 27, 2017
|
|
1QVT
 
 | |
5F7P
 
 | |
5F7R
 
 | |
1OKR
 
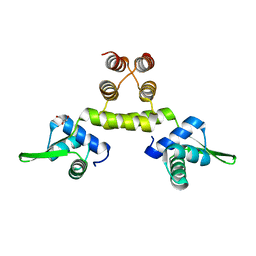 | | Three-dimensional structure of S.aureus methicillin-resistance regulating transcriptional repressor MecI. | | Descriptor: | CHLORIDE ION, GLYCEROL, METHICILLIN RESISTANCE REGULATORY PROTEIN MECI | | Authors: | Garcia-Castellanos, R, Marrero, A, Mallorqui-Fernandez, G, Potempa, J, Coll, M, Gomis-Ruth, F.X. | | Deposit date: | 2003-07-28 | | Release date: | 2003-10-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Three-Dimensional Structure of Meci: Molecular Basis for Transcriptional Regulation of Staphylococcal Methicillin Resistance
J.Biol.Chem., 278, 2003
|
|
5F7Q
 
 | |
