7YLW
 
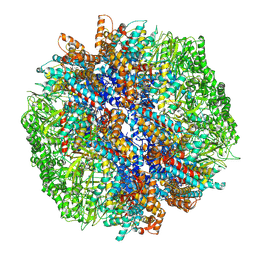 | | yeast TRiC-plp2-tubulin complex at S3 closed TRiC state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Han, W.Y. | | Deposit date: | 2022-07-27 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis of plp2-mediated cytoskeletal protein folding by TRiC/CCT.
Sci Adv, 9, 2023
|
|
2CJM
 
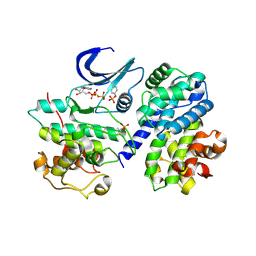 | | Mechanism of CDK inhibition by active site phosphorylation: CDK2 Y15p T160p in complex with cyclin A structure | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Welburn, J.P.I, Tucker, J, Johnson, T, Lindert, L, Morgan, M, Willis, A, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2006-04-05 | | Release date: | 2006-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How Tyrosine 15 Phosphorylation Inhibits the Activity of Cyclin-Dependent Kinase 2-Cyclin A.
J.Biol.Chem., 282, 2007
|
|
2VT5
 
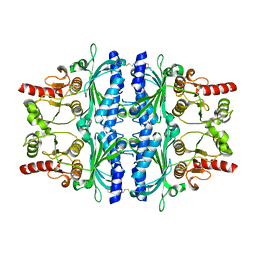 | | FRUCTOSE-1,6-BISPHOSPHATASE(D-FRUCTOSE-1,6-BISPHOSPHATE -1- PHOSPHOHYDROLASE) (E.C.3.1.3.11) COMPLEXED WITH A DUAL BINDING AMP SITE INHIBITOR | | Descriptor: | 4-AMINO-N-[(2-SULFANYLETHYL)CARBAMOYL]BENZENESULFONAMIDE, FRUCTOSE-1,6-BISPHOSPHATASE 1 | | Authors: | Ruf, A, Joseph, C, Benz, J, Fol, B, Tetaz, T. | | Deposit date: | 2008-05-09 | | Release date: | 2008-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Allosteric Fbpase Inhibitors Gain 10(5) Times in Potency When Simultaneously Binding Two Neighboring AMP Sites.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2VUT
 
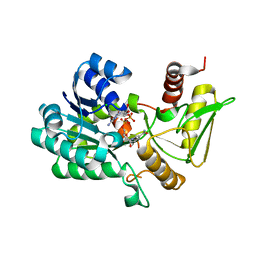 | | Crystal structure of NAD-bound NmrA-AreA zinc finger complex | | Descriptor: | CHLORIDE ION, GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Kotaka, M, Johnson, C, Lamb, H.K, Hawkins, A.R, Ren, J, Stammers, D.K. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of the Recognition of the Negative Regulator Nmra and DNA by the Zinc Finger from the Gata-Type Transcription Factor Area.
J.Mol.Biol., 381, 2008
|
|
2W99
 
 | | Crystal Structure of CDK4 in complex with a D-type cyclin | | Descriptor: | CELL DIVISION PROTEIN KINASE 4, G1/S-SPECIFIC CYCLIN-D1 | | Authors: | Day, P.J, Cleasby, A, Tickle, I.J, Reilly, M.O, Coyle, J.E, Holding, F.P, McMenamin, R.L, Yon, J, Chopra, R, Lengauer, C, Jhoti, H. | | Deposit date: | 2009-01-22 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Human Cdk4 in Complex with a D-Type Cyclin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
5IBK
 
 | | Skp1-F-box in complex with a ubiquitin variant | | Descriptor: | F-box/WD repeat-containing protein 7, Polyubiquitin-B, S-phase kinase-associated protein 1,S-phase kinase-associated protein 1 | | Authors: | Orlicky, S, Sicheri, F. | | Deposit date: | 2016-02-22 | | Release date: | 2016-03-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Inhibition of SCF ubiquitin ligases by engineered ubiquitin variants that target the Cul1 binding site on the Skp1-F-box interface.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7JSL
 
 | |
2AST
 
 | | Crystal structure of Skp1-Skp2-Cks1 in complex with a p27 peptide | | Descriptor: | BENZAMIDINE, Cyclin-dependent kinase inhibitor 1B, Cyclin-dependent kinases regulatory subunit 1, ... | | Authors: | Hao, B, Zhang, N, Schulman, B.A, Wu, G, Pagano, M, Pavletich, N.P. | | Deposit date: | 2005-08-24 | | Release date: | 2005-10-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of the Cks1-Dependent Recognition of p27(Kip1) by the SCF(Skp2) Ubiquitin Ligase.
Mol.Cell, 20, 2005
|
|
7Y67
 
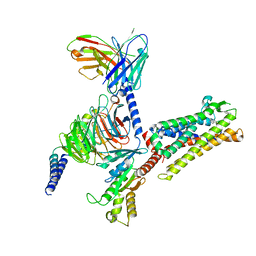 | | Cryo-EM structure of C089-bound C5aR1(I116A) mutant in complex with Gi protein | | Descriptor: | C089 peptide, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y65
 
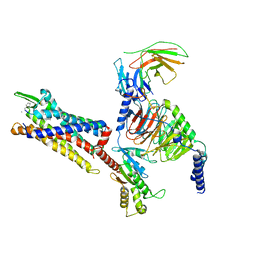 | | Cryo-EM structure of C5a peptide-bound C5aR1 in complex with Gi protein | | Descriptor: | C5a anaphylatoxin chemotactic receptor 1, C5apep peptide, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y66
 
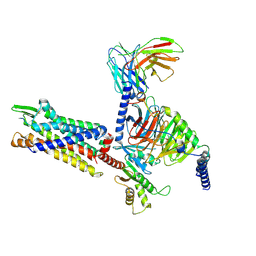 | | Cryo-EM structure of BM213-bound C5aR1 in complex with Gi protein | | Descriptor: | BM213 peptide, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
7Y64
 
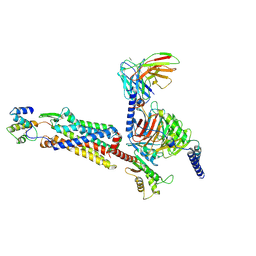 | | Cryo-EM structure of C5a-bound C5aR1 in complex with Gi protein | | Descriptor: | C5a anaphylatoxin, C5a anaphylatoxin chemotactic receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Feng, Y.Y, Zhao, C, Yan, W, Shao, Z.H. | | Deposit date: | 2022-06-18 | | Release date: | 2023-03-01 | | Last modified: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of activation and biased signaling in complement receptor C5aR1.
Cell Res., 33, 2023
|
|
2W9F
 
 | | Crystal Structure of CDK4 in complex with a D-type cyclin | | Descriptor: | CELL DIVISION PROTEIN KINASE 4, G1/S-SPECIFIC CYCLIN-D1 | | Authors: | Day, P.J, Cleasby, A, Tickle, I.J, Reilly, M.O, Coyle, J.E, Holding, F.P, McMenamin, R.L, Yon, J, Chopra, R, Lengauer, C, Jhoti, H. | | Deposit date: | 2009-01-23 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structure of Human Cdk4 in Complex with a D-Type Cyclin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2VUU
 
 | | Crystal structure of NADP-bound NmrA-AreA zinc finger complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Kotaka, M, Johnson, C, Lamb, H.K, Hawkins, A.R, Ren, J, Stammers, D.K. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Analysis of the Recognition of the Negative Regulator Nmra and DNA by the Zinc Finger from the Gata-Type Transcription Factor Area.
J.Mol.Biol., 381, 2008
|
|
6V0O
 
 | | PRMT5 bound to the PBM peptide from pICln | | Descriptor: | ACETYL GROUP, Methylosome protein 50, PBM peptide, ... | | Authors: | McMillan, B.J, Raymond, D.D. | | Deposit date: | 2019-11-19 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Molecular basis for substrate recruitment to the PRMT5 methylosome.
Mol.Cell, 81, 2021
|
|
2VUS
 
 | | Crystal structure of unliganded NmrA-AreA zinc finger complex | | Descriptor: | CHLORIDE ION, NITROGEN METABOLITE REPRESSION REGULATOR NMRA, NITROGEN REGULATORY PROTEIN AREA, ... | | Authors: | Kotaka, M, Johnson, C, Lamb, H.K, Hawkins, A.R, Ren, J, Stammers, D.K. | | Deposit date: | 2008-05-30 | | Release date: | 2008-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of the Recognition of the Negative Regulator Nmra and DNA by the Zinc Finger from the Gata-Type Transcription Factor Area.
J.Mol.Biol., 381, 2008
|
|
6UXX
 
 | | PRMT5:MEP50 Complexed with Allosteric Inhibitor Compound 1a | | Descriptor: | (5R)-2-amino-5-(4-methoxyphenyl)-3-methyl-5-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-yl]-3,5-dihydro-4H-imidazol-4-one, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | Palte, R.L, Schneider, S.E. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Allosteric Modulation of Protein Arginine Methyltransferase 5 (PRMT5).
Acs Med.Chem.Lett., 11, 2020
|
|
7D9V
 
 | |
7D8P
 
 | |
7D8H
 
 | |
6V0P
 
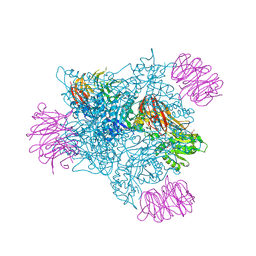 | | PRMT5 complex bound to covalent PBM inhibitor BRD6711 | | Descriptor: | 2-(5-chloro-6-oxopyridazin-1(6H)-yl)-N-(4-methyl-3-sulfamoylphenyl)acetamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | McMillan, B.J, McKinney, D.C. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of a First-in-Class Inhibitor of the PRMT5-Substrate Adaptor Interaction.
J.Med.Chem., 64, 2021
|
|
6IDF
 
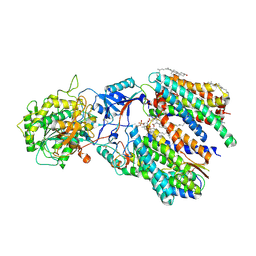 | | Cryo-EM structure of gamma secretase in complex with a Notch fragment | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yang, G, Zhou, R, Zhou, Q, Guo, X, Yan, C, Ke, M, Lei, J, Shi, Y. | | Deposit date: | 2018-09-09 | | Release date: | 2018-12-26 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis of Notch recognition by human gamma-secretase
Nature, 565, 2019
|
|
2W96
 
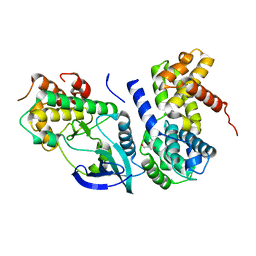 | | Crystal Structure of CDK4 in complex with a D-type cyclin | | Descriptor: | CELL DIVISION PROTEIN KINASE 4, G1/S-SPECIFIC CYCLIN-D1, GLYCEROL | | Authors: | Day, P.J, Cleasby, A, Tickle, I.J, Reilly, M.O, Coyle, J.E, Holding, F.P, McMenamin, R.L, Yon, J, Chopra, R, Lengauer, C, Jhoti, H. | | Deposit date: | 2009-01-21 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Human Cdk4 in Complex with a D-Type Cyclin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6SIO
 
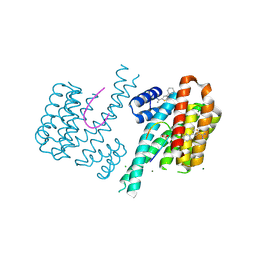 | | Fragment AZ-017 binding at the p53pT387/14-3-3 sigma interface | | Descriptor: | 14-3-3 protein sigma, 5-methyl-4-phenyl-thiophene-2-carboximidamide, CHLORIDE ION, ... | | Authors: | Leysen, S, Wolter, M, Guillory, X, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-08-10 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.60418 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
2B54
 
 | | Human cyclin dependent kinase 2 (CKD2)complexed with DIN-232305 | | Descriptor: | 6-(3,4-DIHYDROXYBENZYL)-3-ETHYL-1-(2,4,6-TRICHLOROPHENYL)-1H-PYRAZOLO[3,4-D]PYRIMIDIN-4(5H)-ONE, Cell division protein kinase 2 | | Authors: | Chang, C.-C. | | Deposit date: | 2005-09-27 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and biological evaluation of 1-aryl-4,5-dihydro-1h-pyraxolo[3,4-d]pyrimidin-4-one inhibitors of cyclin dependent kinases
J.Med.Chem., 47, 2004
|
|
