7YI4
 
 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in close state | | Descriptor: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | Deposit date: | 2022-07-14 | | Release date: | 2023-06-14 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
1ID3
 
 | | CRYSTAL STRUCTURE OF THE YEAST NUCLEOSOME CORE PARTICLE REVEALS FUNDAMENTAL DIFFERENCES IN INTER-NUCLEOSOME INTERACTIONS | | Descriptor: | HISTONE H2A.1, HISTONE H2B.2, HISTONE H3, ... | | Authors: | White, C.L, Suto, R.K, Luger, K. | | Deposit date: | 2001-04-03 | | Release date: | 2001-09-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the yeast nucleosome core particle reveals fundamental changes in internucleosome interactions.
EMBO J., 20, 2001
|
|
7EA8
 
 | |
8G86
 
 | | Human Oct4 bound to nucleosome with human nMatn1 sequence (focused refinement of nucleosome) | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
8G88
 
 | | Human Oct4 bound to nucleosome with human nMatn1 sequence | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
8G8B
 
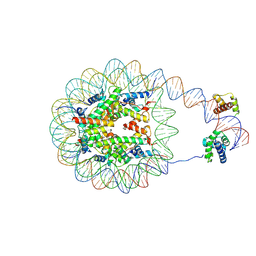 | | Nucleosome with human nMatn1 sequence in complex with Human Oct4 | | Descriptor: | Histone H2A, Histone H2B, Histone H3, ... | | Authors: | Sinha, K.K, Bilokapic, S, Du, Y, Malik, D, Halic, M. | | Deposit date: | 2023-02-17 | | Release date: | 2023-03-22 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Histone modifications regulate pioneer transcription factor cooperativity.
Nature, 619, 2023
|
|
8QZ0
 
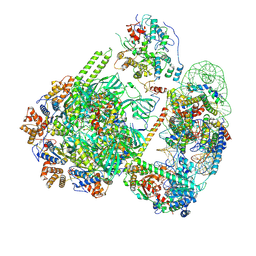 | | SWR1-hexasome-dimer complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2023-10-26 | | Release date: | 2024-10-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Stabilization of the hexasome intermediate during histone exchange by yeast SWR1 complex.
Mol.Cell, 84, 2024
|
|
3FPD
 
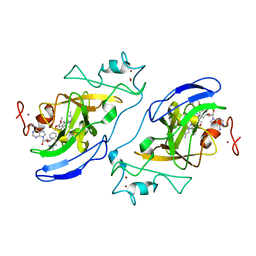 | | G9a-like protein lysine methyltransferase inhibition by BIX-01294 | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-9 specific 5, N-(1-benzylpiperidin-4-yl)-6,7-dimethoxy-2-(4-methyl-1,4-diazepan-1-yl)quinazolin-4-amine, ... | | Authors: | Chang, Y, Zhang, X, Horton, J.R, Cheng, X. | | Deposit date: | 2009-01-05 | | Release date: | 2009-02-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for G9a-like protein lysine methyltransferase inhibition by BIX-01294.
Nat.Struct.Mol.Biol., 16, 2009
|
|
8QKV
 
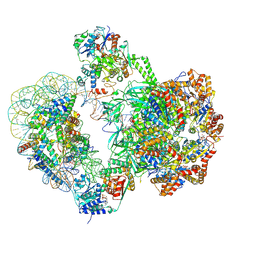 | | SWR1-nucleosome complex in configuration 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2023-09-18 | | Release date: | 2024-10-02 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Nucleosome flipping drives kinetic proofreading and processivity by SWR1.
Nature, 636, 2024
|
|
8QKU
 
 | | SWR1-nucleosome complex in configuration 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2023-09-18 | | Release date: | 2024-10-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Nucleosome flipping drives kinetic proofreading and processivity by SWR1.
Nature, 636, 2024
|
|
8QYV
 
 | | SWR1-hexasome complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-like protein ARP6, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Jalal, A.S.B, Wigley, D.B. | | Deposit date: | 2023-10-26 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Stabilization of the hexasome intermediate during histone exchange by yeast SWR1 complex.
Mol.Cell, 84, 2024
|
|
7UVA
 
 | | Crystal structure of KDM2A histone demethylase catalytic domain in complex with an H3C36 peptide modified by UNC8015 | | Descriptor: | FE (III) ION, Histone H3.2, Lysine-specific demethylase 2A, ... | | Authors: | Budziszewski, G.R, Azzam, D.N, Spangler, C.J, Skrajna, A, Foley, C.A, James, L.I, Frye, S.V, McGinty, R.K. | | Deposit date: | 2022-04-29 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis of paralog-specific KDM2A/B nucleosome recognition.
Nat.Chem.Biol., 19, 2023
|
|
7EA5
 
 | |
2KWK
 
 | | Solution structures of the double PHD fingers of human transcriptional protein DPF3b bound to a H3 peptide wild type | | Descriptor: | Histone peptide, ZINC ION, Zinc finger protein DPF3 | | Authors: | Zeng, L, Zhang, Q, Li, S, Plotnikov, A.N, Walsh, M.J, Zhou, M. | | Deposit date: | 2010-04-12 | | Release date: | 2010-07-14 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Mechanism and regulation of acetylated histone binding by the tandem PHD finger of DPF3b.
Nature, 466, 2010
|
|
6OTC
 
 | | Synthetic Fab bound to Marburg virus VP35 interferon inhibitory domain | | Descriptor: | CHLORIDE ION, GLYCEROL, Polymerase cofactor VP35, ... | | Authors: | Amatya, P, Chen, G, Borek, D, Sidhu, S.S, Leung, D.W. | | Deposit date: | 2019-05-02 | | Release date: | 2019-06-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Marburg Virus RNA Synthesis by a Synthetic Anti-VP35 Antibody.
Acs Infect Dis., 5, 2019
|
|
6PWX
 
 | | Cryo-EM structure of RbBP5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
9B2U
 
 | | Haspin bound to H3 tail | | Descriptor: | Histone H3.2, Serine/threonine-protein kinase haspin | | Authors: | Hicks, C.W, Wolberger, C. | | Deposit date: | 2024-03-16 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Haspin binds to nucleosomes using a unique DNA binding mechanism
To be published
|
|
7K63
 
 | |
7K61
 
 | | Cryo-EM structure of 197bp nucleosome aided by scFv | | Descriptor: | DNA (197-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Zhou, B.-R, Bai, Y. | | Deposit date: | 2020-09-17 | | Release date: | 2020-11-25 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Distinct Structures and Dynamics of Chromatosomes with Different Human Linker Histone Isoforms.
Mol.Cell, 81, 2021
|
|
7K5Y
 
 | |
8U5H
 
 | |
8UXQ
 
 | |
8SR6
 
 | | Crystal structure of legAS4 from Legionella pneumophila subsp. pneumophila with histone H3 (3-17)peptide | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Eukaryotic huntingtin interacting protein B, ... | | Authors: | Xu, C, Chung, I.Y.W, Cygler, M. | | Deposit date: | 2023-05-05 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The SET and ankyrin domains of the secreted Legionella pneumophila histone methyltransferase work together to modify host chromatin.
Mbio, 14, 2023
|
|
6VEN
 
 | |
6G0L
 
 | | Structure of two molecules of the chromatin remodelling enzyme Chd1 bound to a nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chromo domain-containing protein 1, ... | | Authors: | Sundaramoorthy, R, Owen-hughes, T, Norman, D.G, Hughes, A. | | Deposit date: | 2018-03-19 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structure of the chromatin remodelling enzyme Chd1 bound to a ubiquitinylated nucleosome.
Elife, 7, 2018
|
|
