5LVX
 
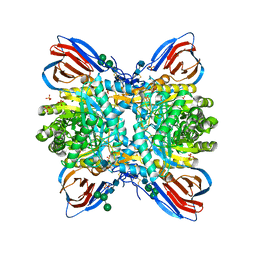 | | Crystal structure of glucocerebrosidase with an inhibitory quinazoline modulator | | Descriptor: | 11-[(2~{R})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 11-[(2~{S})-2-[(2-pyridin-3-ylquinazolin-4-yl)amino]-2,3-dihydro-1~{H}-inden-5-yl]undec-10-ynoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zheng, J, Chen, L, Skinner, O.S, Lansbury, P, Skerlj, R, Mrosek, M, Heunisch, U, Krapp, S, Weigand, S, Charrow, J, Schwake, M, Kelleher, N.L, Silverman, R.B, Krainc, D. | | Deposit date: | 2016-09-14 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | beta-Glucocerebrosidase Modulators Promote Dimerization of beta-Glucocerebrosidase and Reveal an Allosteric Binding Site.
J. Am. Chem. Soc., 140, 2018
|
|
7EA8
 
 | |
5QXK
 
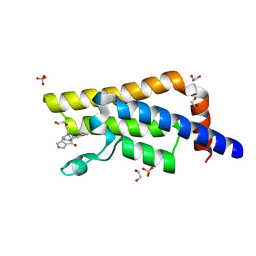 | | PanDDA analysis group deposition -- Crystal Structure of ATAD2 in complex with PC581 | | Descriptor: | (3aS,8S,9aS)-2-acetyl-10-methyl-2,3,6,8,9,9a-hexahydro-3a,8-epiminocyclohepta[1,2-c:4,5-c']dipyrrol-4(1H)-one, 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, ... | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | PanDDA analysis group deposition - Bromodomain of human ATAD2 fragment screening
To Be Published
|
|
1N6F
 
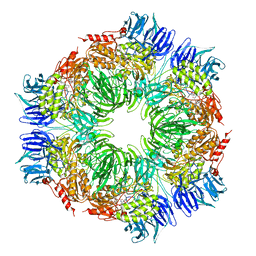 | | tricorn protease in complex with Z-Phe-diketo-Arg-Glu-Phe | | Descriptor: | 4-[2-(3-BENZYLOXYCARBONYLAMINO-4-CYCLOHEXYL-1-HYDROXY-2-OXO-BUTYLAMINO)-5-GUANIDINO-PENTANOYLAMINO]-4-(1-CARBOXY-2-CYCLOHEXYL-ETHYLCARBAMOYL)-BUTYRIC ACID, tricorn protease | | Authors: | Kim, J.-S, Groll, M, Huber, R, Brandstetter, H. | | Deposit date: | 2002-11-10 | | Release date: | 2002-12-30 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Navigation Inside a Protease: Substrate Selection and Product Exit in the Tricorn Protease
from Thermoplasma acidophilum
J.Mol.Biol., 324, 2002
|
|
4O82
 
 | |
2OO8
 
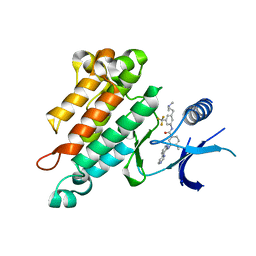 | | Synthesis, Structural Analysis, and SAR Studies of Triazine Derivatives as Potent, Selective Tie-2 Inhibitors | | Descriptor: | Angiopoietin-1 receptor, N-{3-[3-(DIMETHYLAMINO)PROPYL]-5-(TRIFLUOROMETHYL)PHENYL}-4-METHYL-3-[(3-PYRIMIDIN-4-YLPYRIDIN-2-YL)AMINO]BENZAMIDE | | Authors: | Bellon, S.F, Kim, J. | | Deposit date: | 2007-01-25 | | Release date: | 2007-03-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis, structural analysis, and SAR studies of triazine derivatives as potent, selective Tie-2 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3VKF
 
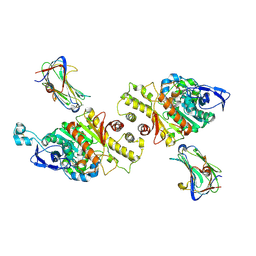 | | Crystal Structure of Neurexin 1beta/Neuroligin 1 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurexin-1-beta, ... | | Authors: | Tanaka, H, Miyazaki, N, Nogi, T, Iwasaki, K, Takagi, J. | | Deposit date: | 2011-11-15 | | Release date: | 2012-08-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Higher-order architecture of cell adhesion mediated by polymorphic synaptic adhesion molecules neurexin and neuroligin.
Cell Rep, 2, 2012
|
|
3P5R
 
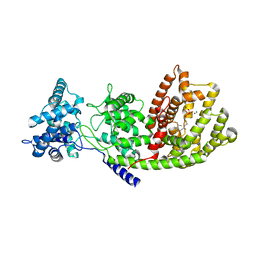 | | Crystal Structure of Taxadiene Synthase from Pacific Yew (Taxus brevifolia) in complex with Mg2+ and 2-fluorogeranylgeranyl diphosphate | | Descriptor: | (2Z,6E,10E)-2-fluoro-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-yl trihydrogen diphosphate, MAGNESIUM ION, Taxadiene synthase | | Authors: | Koksal, M, Christianson, D.W. | | Deposit date: | 2010-10-10 | | Release date: | 2010-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.247 Å) | | Cite: | Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis.
Nature, 469, 2011
|
|
3I7S
 
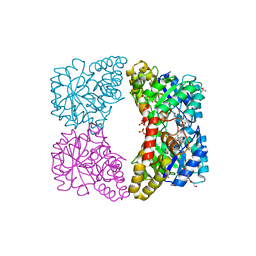 | | Dihydrodipicolinate synthase mutant - K161A - with the substrate pyruvate bound in the active site. | | Descriptor: | Dihydrodipicolinate synthase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Dobson, R.C.J, Jameson, G.B, Gerrard, J.A, Soares da Costa, T.P. | | Deposit date: | 2009-07-08 | | Release date: | 2010-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How essential is the 'essential' active-site lysine in dihydrodipicolinate synthase?
Biochimie, 92, 2010
|
|
5V7T
 
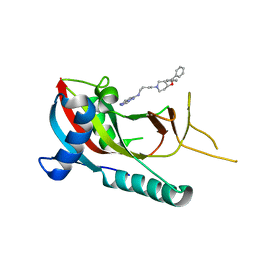 | | crystal structure of PARP14 bound to N-{4-[4-(diphenylmethoxy)piperidin-1-yl]butyl}[1,2,4]triazolo[4,3-b]pyridazin-6-amine inhibitor | | Descriptor: | N-{4-[4-(diphenylmethoxy)piperidin-1-yl]butyl}[1,2,4]triazolo[4,3-b]pyridazin-6-amine, Poly [ADP-ribose] polymerase 14 | | Authors: | saikatendu, k.s, Hirozane, M. | | Deposit date: | 2017-03-20 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification of PARP14 inhibitors using novel methods for detecting auto-ribosylation.
Biochem. Biophys. Res. Commun., 486, 2017
|
|
5EIR
 
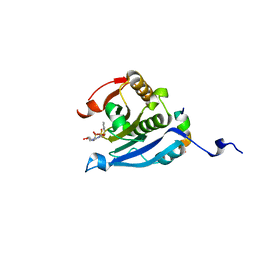 | | Co-crystal structure of eIF4E with nucleotide mimetic inhibitor. | | Descriptor: | Eukaryotic translation initiation factor 4 gamma 1, Eukaryotic translation initiation factor 4E, SULFATE ION, ... | | Authors: | Nowicki, M.W, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2015-10-30 | | Release date: | 2016-09-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design of nucleotide-mimetic and non-nucleotide inhibitors of the translation initiation factor eIF4E: Synthesis, structural and functional characterisation.
Eur.J.Med.Chem., 124, 2016
|
|
3BJ8
 
 | |
1WVX
 
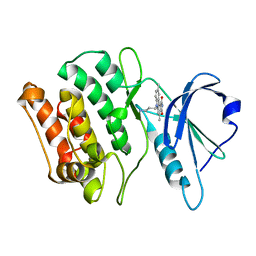 | | Crystal structures of kinase domain of DAP kinase in complex with small molecular inhibitors | | Descriptor: | 6-(3-AMINOPROPYL)-4,9-DIMETHYLPYRROLO[3,4-C]CARBAZOLE-1,3(2H,6H)-DIONE, Death-associated protein kinase 1 | | Authors: | Ueda, Y, Ogata, H, Yamakawa, A, Higuchi, Y. | | Deposit date: | 2004-12-27 | | Release date: | 2006-04-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complex structure of kinase domain of DAP kinase with BDB402
To be Published
|
|
3BJR
 
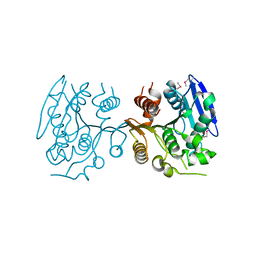 | |
5QXS
 
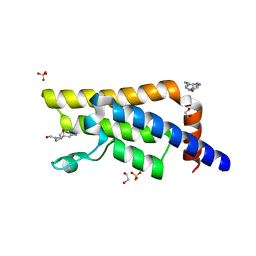 | | PanDDA analysis group deposition -- Crystal Structure of ATAD2 in complex with DF852 | | Descriptor: | (4aR,7aR,9R)-3,4,7,7a,8,9-hexahydro-4a,9-epoxypyrrolo[3',4':4,5]cyclohepta[1,2-d]imidazole-6(5H)-carbaldehyde, 1,2-ETHANEDIOL, ATPase family AAA domain-containing protein 2, ... | | Authors: | Snee, M, Talon, R, Fowley, D, Collins, P, Nelson, A, Arrowsmith, C.H, Bountra, C, Edwards, A, Von-Delft, F. | | Deposit date: | 2020-03-09 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PanDDA analysis group deposition - Bromodomain of human ATAD2 fragment screening
To Be Published
|
|
7JIB
 
 | | Room Temperature Crystal Structure of Nsp10/Nsp16 from SARS-CoV-2 with Substrates and Products of 2'-O-methylation of the Cap-1 | | Descriptor: | 2'-O-methyltransferase, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-(2'-O-METHYL)-ADENOSINE, ... | | Authors: | Wilamowski, M, Minasov, G, Kim, Y, Sherrell, D.A, Shuvalova, L, Lavens, A, Chard, R, Rosas-Lemus, M, Maltseva, N, Jedrzejczak, R, Michalska, K, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-07-23 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2'-O methylation of RNA cap in SARS-CoV-2 captured by serial crystallography.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4RFZ
 
 | |
5ESJ
 
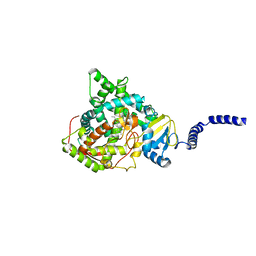 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) G464S mutant complexed with fluconazole | | Descriptor: | 2-(2,4-DIFLUOROPHENYL)-1,3-DI(1H-1,2,4-TRIAZOL-1-YL)PROPAN-2-OL, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R.K, Sabherwal, M, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2015-11-16 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Impact of Homologous Resistance Mutations from Pathogenic Yeast on Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase.
Antimicrob.Agents Chemother., 62, 2018
|
|
3OSF
 
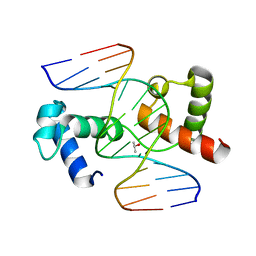 | | The structure of protozoan parasite Trichomonas vaginalis Myb2 in complex with MRE-2f-13 DNA | | Descriptor: | 5'-D(*CP*AP*AP*GP*AP*CP*GP*AP*TP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*AP*TP*CP*GP*TP*CP*TP*TP*G)-3', ISOPROPYL ALCOHOL, ... | | Authors: | Jiang, I, Tsai, C.K, Chen, S.C, Wang, S.H, Amiraslanov, I, Chang, C.F, Wu, W.J, Tai, J.H, Liaw, Y.C, Huang, T.H. | | Deposit date: | 2010-09-09 | | Release date: | 2011-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.032 Å) | | Cite: | Molecular basis of the recognition of the ap65-1 gene transcription promoter elements by a Myb protein from the protozoan parasite Trichomonas vaginalis.
Nucleic Acids Res., 39, 2011
|
|
3BHB
 
 | | Crystal Structure of KMD Phosphopeptide Bound to Human Class I MHC HLA-A2 | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Mohammed, F, Cobbold, M, Zarling, A.L, Salim, M, Barrett-Wilt, G.A, Shabanowitz, J, Hunt, D.F, Engelhard, V.H, Willcox, B.E. | | Deposit date: | 2007-11-28 | | Release date: | 2008-10-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Phosphorylation-dependent interaction between antigenic peptides and MHC class I: a molecular basis for the presentation of transformed self
Nat.Immunol., 9, 2008
|
|
5ESK
 
 | | Saccharomyces cerevisiae CYP51 (Lanosterol 14-alpha demethylase) G464S mutant complexed with itraconazole | | Descriptor: | 2-[(2R)-butan-2-yl]-4-{4-[4-(4-{[(2R,4S)-2-(2,4-dichlorophenyl)-2-(1H-1,2,4-triazol-1-ylmethyl)-1,3-dioxolan-4-yl]methoxy}phenyl)piperazin-1-yl]phenyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, Lanosterol 14-alpha demethylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sagatova, A, Keniya, M.V, Wilson, R.K, Sabherwal, M, Tyndall, J.D.A, Monk, B.C. | | Deposit date: | 2015-11-16 | | Release date: | 2016-11-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Impact of Homologous Resistance Mutations from Pathogenic Yeast on Saccharomyces cerevisiae Lanosterol 14 alpha-Demethylase.
Antimicrob.Agents Chemother., 62, 2018
|
|
6B62
 
 | | IMPase (AF2372) with 400 mM Glutamate | | Descriptor: | Fructose-1,6-bisphosphatase/inositol-1-monophosphatase, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Goldstein, R.I, Roberts, M. | | Deposit date: | 2017-10-01 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | Osmolyte binding capacity of a dual action IMPase/FBPase (AF2372)
To Be Published
|
|
6DJC
 
 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS645 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,N'-(decane-1,10-diyl)bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2018-05-25 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
3B76
 
 | | Crystal structure of the third PDZ domain of human ligand-of-numb protein-X (LNX1) in complex with the C-terminal peptide from the coxsackievirus and adenovirus receptor | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase LNX, SODIUM ION | | Authors: | Ugochukwu, E, Burgess-Brown, N, Berridge, G, Elkins, J, Bunkoczi, G, Pike, A.C.W, Sundstrom, M, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Gileadi, O, von Delft, F, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-30 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the third PDZ domain of human ligand-of-numb protein-X (LNX1) in complex with the C-terminal peptide from the coxsackievirus and adenovirus receptor.
To be Published
|
|
3BIY
 
 | | Crystal structure of p300 histone acetyltransferase domain in complex with a bisubstrate inhibitor, Lys-CoA | | Descriptor: | BROMIDE ION, Histone acetyltransferase p300, [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl (3R,20R)-20-carbamoyl-3-hydroxy-2,2-dimethyl-4,8,14,22-tetraoxo-12-thia-5,9,15,21-tetraazatricos-1-yl dihydrogen diphosphate | | Authors: | Liu, X, Wang, L, Zhao, K, Thompson, P.R, Hwang, Y, Marmorstein, R, Cole, P.A. | | Deposit date: | 2007-12-02 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The structural basis of protein acetylation by the p300/CBP transcriptional coactivator
Nature, 451, 2008
|
|
