4JZW
 
 | |
7KJX
 
 | | Structure of HIV-1 reverse transcriptase initiation complex core with nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 viral RNA fragment, MAGNESIUM ION, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
7KJW
 
 | | Structure of HIV-1 reverse transcriptase initiation complex core with efavirenz | | Descriptor: | (-)-6-CHLORO-4-CYCLOPROPYLETHYNYL-4-TRIFLUOROMETHYL-1,4-DIHYDRO-2H-3,1-BENZOXAZIN-2-ONE, HIV-1 viral RNA fragment, MAGNESIUM ION, ... | | Authors: | Ha, B, Larsen, K.P, Zhang, J, Fu, Z, Montabana, E, Jackson, L.N, Chen, D.H, Puglisi, E.V. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | High-resolution view of HIV-1 reverse transcriptase initiation complexes and inhibition by NNRTI drugs.
Nat Commun, 12, 2021
|
|
3RRX
 
 | | Crystal Structure of Q683A mutant of Exo-1,3/1,4-beta-glucanase (ExoP) from Pseudoalteromonas sp. BB1 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Exo-1,3/1,4-beta-glucanase, ... | | Authors: | Nakatani, Y, Cutfield, S.M, Cutfield, J.F. | | Deposit date: | 2011-05-01 | | Release date: | 2011-12-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and activity of exo-1,3/1,4-beta-glucanase from marine bacterium Pseudoalteromonas sp. BB1 showing a novel C-terminal domain
Febs J., 279, 2012
|
|
1OIM
 
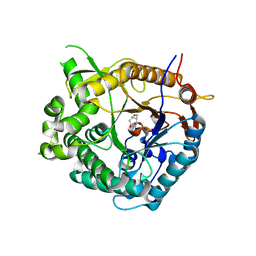 | | Family 1 b-glucosidase from Thermotoga maritima | | Descriptor: | 1-DEOXYNOJIRIMYCIN, BETA-GLUCOSIDASE A | | Authors: | Gloster, T, Zechel, D.L, Boraston, A.B, Boraston, C.M, Macdonald, J.M, Tilbrook, D.M, Stick, R.V, Davies, G.J. | | Deposit date: | 2003-06-19 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Iminosugar Glycosidase Inhibitors: Structural and Thermodynamic Dissection of the Binding of Isofagomine and 1-Deoxynojirimycin to Beta-Glucosidases
J.Am.Chem.Soc., 125, 2003
|
|
4KWP
 
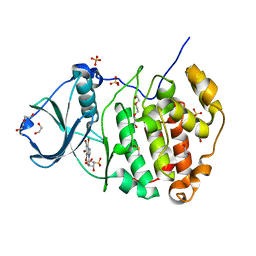 | | Crystal Structure of Human CK2-alpha in complex with a benzimidazole inhibitor (K164) at 1.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 4,5,6,7-tetrabromo-1-(2-deoxy-beta-D-erythro-pentofuranosyl)-1H-benzimidazole, Casein kinase II subunit alpha, ... | | Authors: | Ranchio, A, Lolli, G, Battistutta, R. | | Deposit date: | 2013-05-24 | | Release date: | 2014-04-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Cell-permeable dual inhibitors of protein kinases CK2 and PIM-1: structural features and pharmacological potential.
Cell.Mol.Life Sci., 71, 2014
|
|
4KO0
 
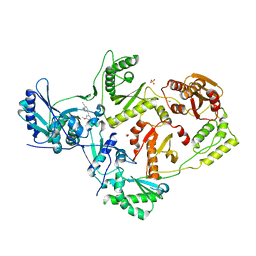 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH an anilinylpyrimidine derivative (JLJ-135) | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-methoxypyrimidin-2-yl)amino]-2-[(3-methylbut-2-en-1-yl)oxy]benzonitrile, HIV-1 reverse transcriptase, ... | | Authors: | Das, K, Bauman, J.D, Arnold, E. | | Deposit date: | 2013-05-10 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Extension into the entrance channel of HIV-1 reverse transcriptase-Crystallography and enhanced solubility.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1KI6
 
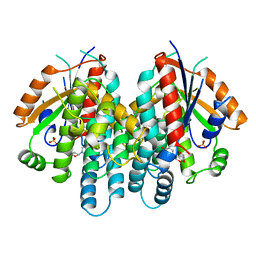 | | CRYSTAL STRUCTURE OF THYMIDINE KINASE FROM HERPES SIMPLEX VIRUS TYPE I COMPLEXED WITH A 5-IODOURACIL ANHYDROHEXITOL NUCLEOSIDE | | Descriptor: | 1',5'-ANHYDRO-2',3'-DIDEOXY-2'-(5-IODOURACIL-1-YL)-D-ABABINO-HEXITOL, SULFATE ION, THYMIDINE KINASE | | Authors: | Champness, J.N, Bennett, M.S, Wien, F, Herdewijn, P, Ostrowski, T, Summers, W.C, Sanderson, M.R. | | Deposit date: | 1998-05-18 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Exploring the active site of herpes simplex virus type-1 thymidine kinase by X-ray crystallography of complexes with aciclovir and other ligands.
Proteins, 32, 1998
|
|
4H4M
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with (E)-3-(3-chloro-5-(4-chloro-2-(2-(2,4-dioxo-3,4- dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)phenyl)acrylonitrile (JLJ494), a Non-nucleoside Inhibitor | | Descriptor: | (2E)-3-(3-chloro-5-{4-chloro-2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}phenyl)prop-2-enenitrile, 1,2-ETHANEDIOL, Reverse transcriptase/ribonuclease H, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2012-09-17 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase with Picomolar Inhibitors Reveal Key Interactions for Drug Design.
J.Am.Chem.Soc., 134, 2012
|
|
3D1R
 
 | | Structure of E. coli GlpX with its substrate fructose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Singer, A, Skarina, T, Dong, A, Brown, G, Joachimiak, A, Edwards, A.M, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-06 | | Release date: | 2008-12-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Biochemical Characterization of the Type II Fructose-1,6-bisphosphatase GlpX from Escherichia coli.
J.Biol.Chem., 284, 2009
|
|
1HNV
 
 | | STRUCTURE OF HIV-1 RT(SLASH)TIBO R 86183 COMPLEX REVEALS SIMILARITY IN THE BINDING OF DIVERSE NONNUCLEOSIDE INHIBITORS | | Descriptor: | 5-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66) | | Authors: | Das, K, Ding, J, Arnold, E. | | Deposit date: | 1995-03-30 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of HIV-1 RT/TIBO R 86183 complex reveals similarity in the binding of diverse nonnucleoside inhibitors.
Nat.Struct.Biol., 2, 1995
|
|
1KLM
 
 | | HIV-1 REVERSE TRANSCRIPTASE COMPLEXED WITH BHAP U-90152 | | Descriptor: | (1-(5-METHANSULPHONAMIDO-1H-INDOL-2-YL-CARBONYL)4-[METHYLAMINO)PYRIDINYL]PIPERAZINE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R.M, Stammers, D.K, Stuart, D.I. | | Deposit date: | 1997-03-17 | | Release date: | 1998-03-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Unique features in the structure of the complex between HIV-1 reverse transcriptase and the bis(heteroaryl)piperazine (BHAP) U-90152 explain resistance mutations for this nonnucleoside inhibitor.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
4LAJ
 
 | | Crystal structure of HIV-1 YU2 envelope gp120 glycoprotein in complex with CD4-mimetic miniprotein, M48U1, and llama single-domain, broadly neutralizing, co-receptor binding site antibody, JM4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6-AMINOHEXANOIC ACID, ... | | Authors: | Acharya, P, Luongo, T.S, Kwong, P.D. | | Deposit date: | 2013-06-20 | | Release date: | 2013-08-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Heavy Chain-Only IgG2b Llama Antibody Effects Near-Pan HIV-1 Neutralization by Recognizing a CD4-Induced Epitope That Includes Elements of Coreceptor- and CD4-Binding Sites.
J.Virol., 87, 2013
|
|
4H4O
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase (RT) in Complex with (E)-3-(3-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)- 4-fluorophenoxy)-5-fluorophenyl)acrylonitrile (JLJ506), A Non-nucleoside inhibitor | | Descriptor: | (2E)-3-(3-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]-4-fluorophenoxy}-5-fluorophenyl)prop-2-enenitrile, Reverse transcriptase/ribonuclease H, Exoribonuclease H, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2012-09-17 | | Release date: | 2012-12-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structures of HIV-1 Reverse Transcriptase with Picomolar Inhibitors Reveal Key Interactions for Drug Design.
J.Am.Chem.Soc., 134, 2012
|
|
1PMJ
 
 | | Crystal structure of Caldicellulosiruptor saccharolyticus CBM27-1 | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, CALCIUM ION, ... | | Authors: | Roske, Y, Sunna, A, Heinemann, U. | | Deposit date: | 2003-06-11 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | High-resolution crystal structures of Caldicellulosiruptor strain Rt8B.4 carbohydrate-binding module CBM27-1 and its complex with mannohexaose.
J.Mol.Biol., 340, 2004
|
|
3T2C
 
 | | Fructose-1,6-bisphosphate aldolase/phosphatase from Thermoproteus neutrophilus, DHAP-bound form | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-1,6-bisphosphate aldolase/phosphatase, MAGNESIUM ION | | Authors: | Du, J, Say, R, Lue, W, Fuchs, G, Einsle, O. | | Deposit date: | 2011-07-22 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Active-site remodelling in the bifunctional fructose-1,6-bisphosphate aldolase/phosphatase.
Nature, 478, 2011
|
|
4EPJ
 
 | | Crystal Structure of inactive single chain wild-type HIV-1 Protease in Complex with the substrate p2-NC | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-04-17 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural, kinetic, and thermodynamic studies of specificity designed HIV-1 protease.
Protein Sci., 21, 2012
|
|
4EQJ
 
 | | Crystal Structure of inactive single chain variant of HIV-1 Protease in Complex with the substrate RT-RH | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, protease, ... | | Authors: | Schiffer, C.A, Mittal, S. | | Deposit date: | 2012-04-19 | | Release date: | 2012-06-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, kinetic, and thermodynamic studies of specificity designed HIV-1 protease.
Protein Sci., 21, 2012
|
|
4ICL
 
 | | HIV-1 reverse transcriptase with bound fragment at the incoming dNTP binding site | | Descriptor: | 4-(4-methylpiperazin-1-yl)benzoic acid, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-10 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4IG0
 
 | | HIV-1 reverse transcriptase with bound fragment at the 507 site | | Descriptor: | 2-({[2-(3,4-dihydroquinolin-1(2H)-yl)-2-oxoethyl](methyl)amino}methyl)quinazolin-4(1H)-one, DIMETHYL SULFOXIDE, P51 RT, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
1M8S
 
 | | Crystal Structures of Cadmium-binding Acidic Phospholipase A2 from the Venom of Agkistrodon halys pallas at 1.9 Resolution (crystal grown at pH 5.9) | | Descriptor: | 1,4-BUTANEDIOL, CADMIUM ION, phospholipase a2 | | Authors: | Xu, S, Gu, L, Zhou, Y, Lin, Z. | | Deposit date: | 2002-07-25 | | Release date: | 2003-02-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of cadmium-binding acidic phospholipase A(2) from the venom of Agkistrodon halys Pallas at 1.9A resolutio
Biochem.Biophys.Res.Commun., 300, 2003
|
|
1GVF
 
 | | Structure of tagatose-1,6-bisphosphate aldolase | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHOGLYCOLOHYDROXAMIC ACID, SODIUM ION, ... | | Authors: | Hall, D.R, Hunter, W.N. | | Deposit date: | 2002-02-11 | | Release date: | 2002-06-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of Tagatose-1,6-Bisphosphate Aldolase; Insight Into Chiral Discrimination, Mechanism and Specificity of Class II Aldolases
J.Biol.Chem., 277, 2002
|
|
5NPF
 
 | | Crystal structure of txGH116 (beta-glucosidase from Thermoanaerobacterium xylolyticum) in complex with beta Cyclophellitol Cyclosulfate probe ME594 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Glucosylceramidase, ... | | Authors: | Wu, L, Offen, W.A, Breen, I.Z, Davies, G.J. | | Deposit date: | 2017-04-16 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | 1,6-Cyclophellitol Cyclosulfates: A New Class of Irreversible Glycosidase Inhibitor.
ACS Cent Sci, 3, 2017
|
|
2Q8M
 
 | | T-like Fructose-1,6-bisphosphatase from Escherichia coli with AMP, Glucose 6-phosphate, and Fructose 1,6-bisphosphate bound | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Hines, J.K, Kruesel, C.E, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2007-06-11 | | Release date: | 2007-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Inhibited Fructose-1,6-bisphosphatase from Escherichia coli: DISTINCT ALLOSTERIC INHIBITION SITES FOR AMP AND GLUCOSE 6-PHOSPHATE AND THE CHARACTERIZATION OF A GLUCONEOGENIC SWITCH.
J.Biol.Chem., 282, 2007
|
|
1W8S
 
 | | The mechanism of the Schiff Base Forming Fructose-1,6-bisphosphate Aldolase: Structural analysis of reaction intermediates | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, FRUCTOSE-BISPHOSPHATE ALDOLASE CLASS I | | Authors: | Lorentzen, E, Hensel, R, Siebers, B, Pohl, E. | | Deposit date: | 2004-09-27 | | Release date: | 2005-03-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of the Schiff base forming fructose-1,6-bisphosphate aldolase: structural analysis of reaction intermediates.
Biochemistry, 44, 2005
|
|
