8VK2
 
 | | X-ray crystal structure of human IgE 4C8 Fab | | Descriptor: | IgE 4C8 heavy chain, IgE 4C8 light chain | | Authors: | Khatri, K, Ball, A, Smith, S.A, Champan, M.D, Pomes, A, Chruszcz, M. | | Deposit date: | 2024-01-08 | | Release date: | 2024-06-19 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Structural analysis of human IgE monoclonal antibody epitopes on dust mite allergen Der p 2.
J.Allergy Clin.Immunol., 154, 2024
|
|
6XM2
 
 | | The structure of the 4A11.v7 antibody in complex with human TGFb2 | | Descriptor: | 4A11.v7 heavy chain Fab (VH-CH1) IgG1 humanized, 4A11.v7 kappa light chain Fab (VL-CL) humanized, Transforming growth factor beta-2 | | Authors: | Lupardus, P.J, Yin, J.P. | | Deposit date: | 2020-06-29 | | Release date: | 2021-07-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | TGF beta 2 and TGF beta 3 isoforms drive fibrotic disease pathogenesis.
Sci Transl Med, 13, 2021
|
|
6N5E
 
 | |
2RGS
 
 | | FC-fragment of monoclonal antibody IGG2B from Mus musculus | | Descriptor: | Ig gamma-2B heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kolenko, P, Duskova, J, Skalova, T, Dohnalek, J, Hasek, J. | | Deposit date: | 2007-10-05 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | New insights into intra- and intermolecular interactions of immunoglobulins: crystal structure of mouse IgG2b-Fc at 2.1-A resolution
Immunology, 126, 2008
|
|
1L6X
 
 | | FC FRAGMENT OF RITUXIMAB BOUND TO A MINIMIZED VERSION OF THE B-DOMAIN FROM PROTEIN A CALLED Z34C | | Descriptor: | IMMUNOGLOBULIN GAMMA-1 HEAVY CHAIN CONSTANT REGION, Minimized B-domain of Protein A Z34C, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Idusogie, E.E, Presta, L.G, Santoro-Gazzano, H, Totpal, K, Wong, P.Y, Ultsch, M, Meng, Y.G, Mullkerrin, M.G. | | Deposit date: | 2002-03-14 | | Release date: | 2002-04-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mapping of the C1q binding site on rituxan, a chimeric antibody with a human IgG1 Fc.
J.Immunol., 164, 2000
|
|
7TXZ
 
 | |
7TY0
 
 | |
5NLU
 
 | | Structure of Nb36 crystal form 1 | | Descriptor: | SULFATE ION, single domain llama antibody Nb36 | | Authors: | Hansen, S.B, Andersen, K.R, Laursen, N.S, Andersen, G.R. | | Deposit date: | 2017-04-05 | | Release date: | 2017-05-31 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Introducing site-specific cysteines into nanobodies for mercury labelling allows de novo phasing of their crystal structures.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7JV2
 
 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody Fab fragment (local refinement of the receptor-binding motif and Fab variable domains) | | Descriptor: | S2H13 Fab heavy chain, S2H13 Fab light chain, Spike glycoprotein | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
6OY4
 
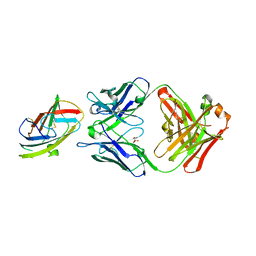 | | Crystal structure of complex between recombinant Der p 2.0103 and Fab fragment of 7A1 | | Descriptor: | Der p 2 variant 3, Fab fragment of IgG, HEAVY CHAIN, ... | | Authors: | Kapingidza, A.B, Offermann, L.R, Glesner, J, Wunschmann, S, Vailes, L.D, Chapman, M.D.C, Pomes, A, Chruszcz, M. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Human IgE Antibody Binding Site on Der p 2 for the Design of a Recombinant Allergen for Immunotherapy.
J Immunol., 203, 2019
|
|
6W41
 
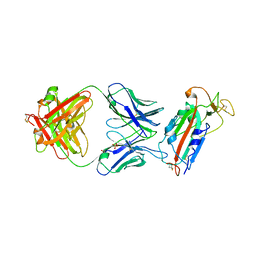 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody CR3022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CR3022 Fab heavy chain, CR3022 Fab light chain, ... | | Authors: | Yuan, M, Wu, N.C, Zhu, X.Y, Wilson, I.A. | | Deposit date: | 2020-03-09 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.084 Å) | | Cite: | A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV.
Science, 368, 2020
|
|
6OHG
 
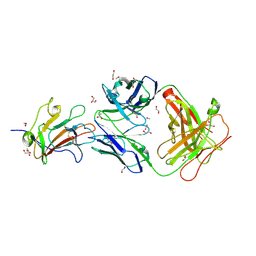 | | Structure of Plasmodium falciparum vaccine candidate Pfs230D1M in complex with the Fab of a transmission blocking antibody | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 4F12 Heavy chain, ... | | Authors: | Garboczi, D.N, Singh, K, Gittis, A.G. | | Deposit date: | 2019-04-05 | | Release date: | 2020-06-17 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Structure and function of a malaria transmission blocking vaccine targeting Pfs230 and Pfs230-Pfs48/45 proteins.
Commun Biol, 3, 2020
|
|
7JV6
 
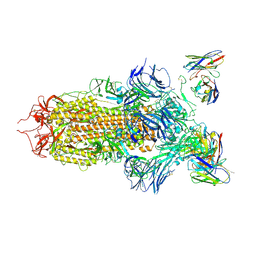 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody (closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2H13 Fab heavy chain, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JV4
 
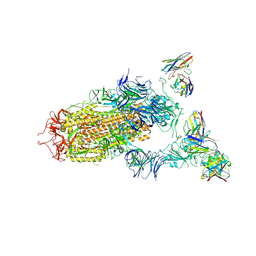 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody (one RBD open) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2H13 Fab heavy chain, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JVA
 
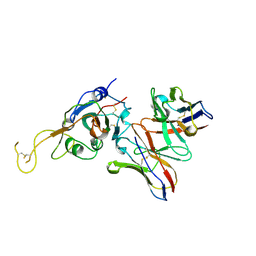 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment (local refinement of the receptor-binding domain and Fab variable domains) | | Descriptor: | S2A4 Fab heavy chain, S2A4 Fab light chain, Spike glycoprotein, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7YZJ
 
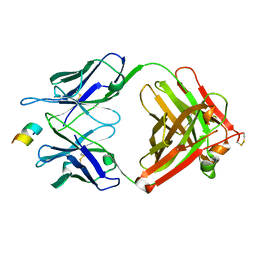 | | FAB IN COMPLEX WITH ANTIGENIC PEPTIDE OF INTERLEUKIN-2 | | Descriptor: | Antigenic peptide, Heavy chain of FAB fragment, Light chain of FAB fragment | | Authors: | Pletnev, V, Pletneva, N. | | Deposit date: | 2022-02-20 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-Dimensional Structure of Fab Fragment of Monoclonal Antibody LNKB-2 Complexed with Antigenic Nonapeptide from Human Interleukin-2
Russ.J.Bioorganic Chem., 2023
|
|
8ASY
 
 | | SARS-CoV-2 Omicron BA.2.75 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-08-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A delicate balance between antibody evasion and ACE2 affinity for Omicron BA.2.75.
Cell Rep, 42, 2022
|
|
1OHQ
 
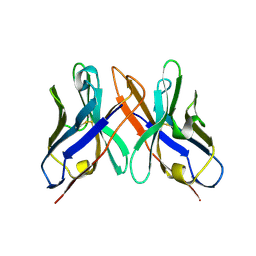 | | Crystal structure of HEL4, a soluble human VH antibody domain resistant to aggregation | | Descriptor: | IMMUNOGLOBULIN | | Authors: | Jespers, L, Schon, O, James, L.C, Veprintsev, D, Winter, G. | | Deposit date: | 2003-05-30 | | Release date: | 2004-03-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Hel4, a Soluble, Refoldable Human V(H) Single Domain with a Germ-Line Scaffold
J.Mol.Biol., 337, 2004
|
|
5UBY
 
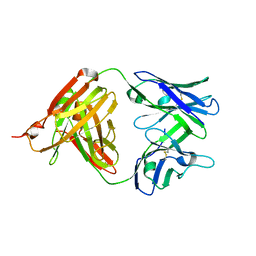 | | Fab structure of anti-HIV-1 gp120 mAb 1A8 | | Descriptor: | Heavy chain of Fab fragment of anti-HIV1 gp120 mAb 1A8, Light chain of Fab fragment of anti-HIV1 gp120 mAb 1A8 | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High Antibody Diversity and Low Inter-clonal Competition Favor Production of Functional Neutralizing Antibodies
To be Published
|
|
7S1B
 
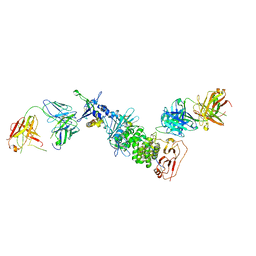 | | Crystal structure of Epstein-Barr virus glycoproteins gH/gL/gp42-peptide in complex with human neutralizing antibodies 769C2 and 770F7 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769C2 Fab heavy chain, 769C2 Fab light chain, ... | | Authors: | Chen, W.-H, Cohen, J.I, Kanekiyo, M, Joyce, M.G. | | Deposit date: | 2021-09-02 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
7S07
 
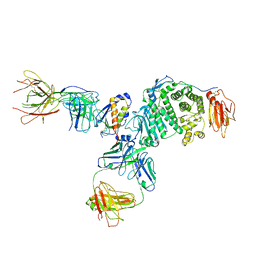 | | Crystal structure of Epstein-Barr virus glycoprotein gH/gL/gp42-peptide in complex with human neutralizing antibodies 769B10 and 769C2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 769B10 Fab heavy chain, 769B10 Fab light chain, ... | | Authors: | Chen, W.-H, Kanekiyo, M, Cohen, J.I, Joyce, M.G. | | Deposit date: | 2021-08-30 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Epstein-Barr virus gH/gL has multiple sites of vulnerability for virus neutralization and fusion inhibition.
Immunity, 55, 2022
|
|
5JXE
 
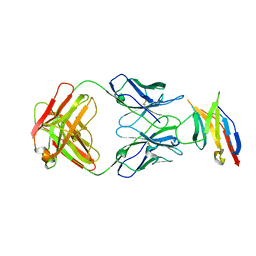 | | Human PD-1 ectodomain complexed with Pembrolizumab Fab | | Descriptor: | Pembrolizumab Fab heavy chain, Pembrolizumab Fab light chain, Programmed cell death protein 1 | | Authors: | Na, Z, Bharath, S.R, Song, H. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for blocking PD-1-mediated immune suppression by therapeutic antibody pembrolizumab.
Cell Res., 27, 2017
|
|
6JC2
 
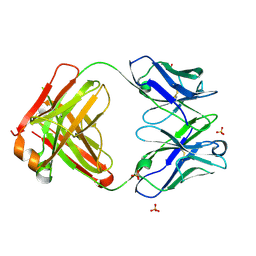 | | Crystal structure of the Fab fragment of ipilimumab | | Descriptor: | SULFATE ION, ipilimumab fab heavy chain, ipilimumab fab light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2019-01-27 | | Release date: | 2019-09-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal Structure of the Fab Fragment of an Anti-CTLA-4 Antibody, Ipilimumab, Used for Cancer Immunotherapy
Bull.Korean Chem.Soc., 40, 2019
|
|
5UBZ
 
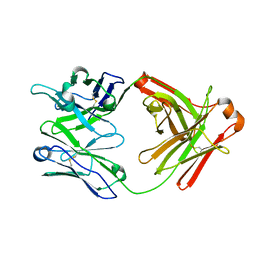 | | Fab structure of HIV gp120 specific mAb 1E12 | | Descriptor: | Anti-HIV1 gp120 mAb 1E12 Fab heavy chain, Anti-HIV1 gp120 mAb 1E12 Fab light chain | | Authors: | Pan, R, Kong, X.-P. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | High Antibody Diversity and Low Inter-clonal Competition Favor Production of Functional Neutralizing Antibodies
To be published
|
|
6VOS
 
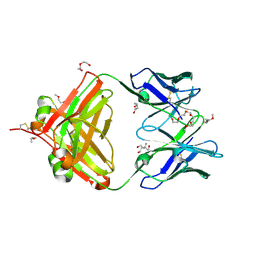 | | Crystal structure of macaque anti-HIV-1 antibody RM20J | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, HEXAETHYLENE GLYCOL, NONAETHYLENE GLYCOL, ... | | Authors: | Yuan, M, Wilson, I.A. | | Deposit date: | 2020-01-31 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
