5C7D
 
 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 17 | | Descriptor: | (2R)-4-[2-(6-chloro-2,3-dihydro-1H-pyrrolo[3,2-c]pyridin-1-yl)-2-oxoethyl]-2-methylpiperazin-1-ium, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-24 | | Release date: | 2015-08-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
5SOU
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000285507655 - (R) and (S) isomers | | Descriptor: | 5-[(3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]-1,3,4-oxadiazol-2(3H)-one, 5-[(3S)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]-1,3,4-oxadiazol-2(3H)-one, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6VJB
 
 | |
5SOT
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with ZINC000292637864 - (R) and (S) isomers | | Descriptor: | Non-structural protein 3, {1-[(3R)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]-1H-1,2,3-triazol-4-yl}methanol, {1-[(3S)-1-(7H-pyrrolo[2,3-d]pyrimidin-4-yl)piperidin-3-yl]-1H-1,2,3-triazol-4-yl}methanol | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5XKR
 
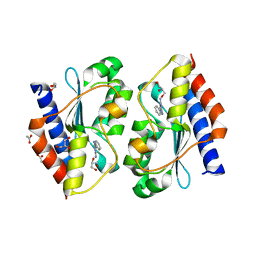 | | Crystal structure of Msmeg3575 in complex with benzoguanamine | | Descriptor: | 6-phenyl-1,3,5-triazine-2,4-diamine, ACETATE ION, CMP/dCMP deaminase, ... | | Authors: | Gaded, V.M, Anand, R. | | Deposit date: | 2017-05-09 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Selective Deamination of Mutagens by a Mycobacterial Enzyme
J. Am. Chem. Soc., 139, 2017
|
|
6CKP
 
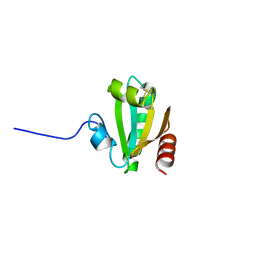 | |
2YNE
 
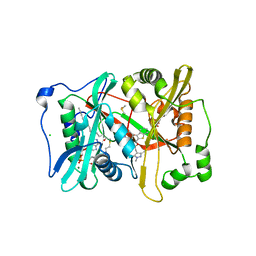 | | Plasmodium vivax N-myristoyltransferase in complex with a benzothiophene inhibitor | | Descriptor: | 2-(3-piperidin-4-yloxy-1-benzothiophen-2-yl)-5-[(1,3,5-trimethylpyrazol-4-yl)methyl]-1,3,4-oxadiazole, 2-oxopentadecyl-CoA, CHLORIDE ION, ... | | Authors: | Wright, M.H, Clough, B, Rackham, M.D, Brannigan, J.A, Grainger, M, Bottrill, A.R, Heal, W.P, Broncel, M, Serwa, R.A, Mann, D, Leatherbarrow, R.J, Wilkinson, A.J, Holder, A.A, Tate, E.W. | | Deposit date: | 2012-10-13 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Validation of N-Myristoyltransferase as an Antimalarial Drug Target Using an Integrated Chemical Biology Approach.
Nat.Chem., 6, 2014
|
|
4QMX
 
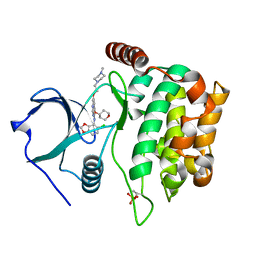 | | MST3 in complex with SARACATINIB | | Descriptor: | N-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-7-[2-(4-METHYLPIPERAZIN-1-YL)ETHOXY]-5-(TETRAHYDRO-2H-PYRAN-4-YLOXY)QUINAZOLIN-4-AMINE, SERINE/THREONINE-PROTEIN KINASE 24 | | Authors: | Olesen, S.H, Watts, C, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2014-06-16 | | Release date: | 2015-07-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Discovery of Diverse Small-Molecule Inhibitors of Mammalian Sterile20-like Kinase 3 (MST3).
Chemmedchem, 11, 2016
|
|
6CLY
 
 | |
5A4L
 
 | | DYRK1A IN COMPLEX WITH FLUORO BENZOTHIAZOLE FRAGMENT | | Descriptor: | DUAL SPECIFICITY TYROSINE-PHOSPHORYLATION- REGULATED KINASE 1A, N-(5-FLUORANYL-1,3-BENZOTHIAZOL-2-YL)ETHANAMIDE | | Authors: | Rothweiler, U. | | Deposit date: | 2015-06-10 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Probing the ATP-Binding Pocket of Protein Kinase Dyrk1A with Benzothiazole Fragment Molecules
J.Med.Chem., 59, 2016
|
|
2Y9G
 
 | | High-resolution Structural Insights on the Sugar-recognition and Fusion Tag Properties of a Versatile b-Trefoil Lectin Domain | | Descriptor: | GLYCEROL, HEMOLYTIC LECTIN LSLA, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Angulo, I, Acebron, I, de las Rivas, B, Munoz, R, Rodriguez, J.I, Menendez, M, Garcia, P, Tateno, H, Goldstein, I.J, Perez-Agote, B, Mancheno, J.M. | | Deposit date: | 2011-02-14 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | High-Resolution Structural Insights on the Sugar-Recognition and Fusion Tag Properties of a Versatile Beta-Trefoil Lectin Domain from the Mushroom Laetiporus Sulphureus.
Glycobiology, 21, 2011
|
|
2CQT
 
 | | Crystal Structure of Cellvibrio gilvus Cellobiose Phosphorylase Crystallized from Sodium/Potassium Phosphate | | Descriptor: | Cellobiose Phosphorylase, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Hidaka, M, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2005-05-20 | | Release date: | 2006-05-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural dissection of the reaction mechanism of cellobiose phosphorylase.
Biochem.J., 398, 2006
|
|
4ZTS
 
 | | Human Aurora A catalytic domain bound to FK1142 | | Descriptor: | (2Z,5Z)-2-[(4-ethylphenyl)imino]-3-methyl-5-[(2-{[4-(1H-tetrazol-5-yl)phenyl]amino}pyridin-4-yl)methylidene]-1,3-thiazolidin-4-one, (4S)-2-METHYL-2,4-PENTANEDIOL, Aurora kinase A | | Authors: | Marcaida, M.J, Kilchmann, F, Schick, T, Reymond, J.L. | | Deposit date: | 2015-05-15 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery of a Selective Aurora A Kinase Inhibitor by Virtual Screening.
J.Med.Chem., 59, 2016
|
|
6VJJ
 
 | | Crystal Structure of wild-type KRAS4b (GMPPNP-bound) in complex with RAS-binding domain (RBD) of RAF1/CRAF | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2020-01-16 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | KRAS interaction with RAF1 RAS-binding domain and cysteine-rich domain provides insights into RAS-mediated RAF activation.
Nat Commun, 12, 2021
|
|
6K9G
 
 | | Human LXR-beta in complex with an agonist | | Descriptor: | Oxysterols receptor LXR-beta, ~{tert}-butyl (2'~{R},3~{R})-2'-[3-[4-(hydroxymethyl)-3-methylsulfonyl-phenyl]phenyl]-2-oxidanylidene-spiro[1~{H}-indole-3,3'-pyrrolidine]-1'-carboxylate | | Authors: | Zhang, Z, Zhou, H. | | Deposit date: | 2019-06-15 | | Release date: | 2020-04-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of new LXR beta agonists as glioblastoma inhibitors.
Eur.J.Med.Chem., 194, 2020
|
|
6G8R
 
 | | SP140 PHD-Bromodomain complex with scFv | | Descriptor: | 1,2-ETHANEDIOL, Nuclear body protein SP140, ZINC ION, ... | | Authors: | Fairhead, M, Graslund, S, Strain-Damerell, C, Picaud, S.S, Pike, A.C.W, Pinkas, D.M, Wigren, E, Preger, C, Persson Lotsholm, H, Ossipova, E, Filippakopoulos, P, Burgess-Brown, N.A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-04-09 | | Release date: | 2018-04-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | SP140 PHD-Bromodomain complex with scFv
To Be Published
|
|
5SPC
 
 | | PanDDA analysis group deposition -- Crystal structure of SARS-CoV-2 NSP3 macrodomain in complex with Z5010894387 - (R,R) and (S,S) isomers | | Descriptor: | (1R,2R)-4-hydroxy-1-[(5,6,7,8-tetrahydro-1,8-naphthyridine-3-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, (1S,2S)-4-hydroxy-1-[(5,6,7,8-tetrahydro-1,8-naphthyridine-3-carbonyl)amino]-2,3-dihydro-1H-indene-2-carboxylic acid, Non-structural protein 3 | | Authors: | Correy, G.J, Fraser, J.S. | | Deposit date: | 2022-06-09 | | Release date: | 2022-07-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Iterative computational design and crystallographic screening identifies potent inhibitors targeting the Nsp3 macrodomain of SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6G0O
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with an acetylated ATRX peptide (K1030ac/K1033ac) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, Transcriptional regulator ATRX | | Authors: | Filippakopoulos, P, Picaud, S, Newman, J, Sorrell, F, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-03-19 | | Release date: | 2018-11-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Interactome Rewiring Following Pharmacological Targeting of BET Bromodomains.
Mol. Cell, 73, 2019
|
|
4ZX9
 
 | |
7GHA
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with DAN-LON-a5fc619e-8 (Mpro-x3077) | | Descriptor: | 1-{(1S,4S)-5-[(3-chlorophenyl)methyl]-2,5-diazabicyclo[2.2.1]heptan-2-yl}ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5A2F
 
 | | Two membrane distal IgSF domains of CD166 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CD166 ANTIGEN | | Authors: | Chappell, P.E, Johnson, S, Lea, S.M, Brown, M.H. | | Deposit date: | 2015-05-18 | | Release date: | 2015-07-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of Cd6 and its Ligand Cd166 Give Insight Into Their Interaction.
Structure, 23, 2015
|
|
5SEJ
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c4(cn1c(nc(n1)CCc2nc(nn2C)N3CCCC3)cc4)Cl, micromolar IC50=0.062679 | | Descriptor: | (4R)-6-chloro-2-{2-[1-methyl-3-(pyrrolidin-1-yl)-1H-1,2,4-triazol-5-yl]ethyl}[1,2,4]triazolo[1,5-a]pyridine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Lerner, C, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
5SEQ
 
 | | CRYSTAL STRUCTURE OF HUMAN PHOSPHODIESTERASE 10 IN COMPLEX WITH c1(nn(c(n1)CCc2nn3c(n2)c(nc(c3C)C)C)C)N4CCCC4, micromolar IC50=0.037262 | | Descriptor: | (4S)-5,6,8-trimethyl-2-{2-[1-methyl-3-(pyrrolidin-1-yl)-1H-1,2,4-triazol-5-yl]ethyl}[1,2,4]triazolo[1,5-a]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Lerner, C, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A high quality, industrial data set for binding affinity prediction: performance comparison in different early drug discovery scenarios.
J.Comput.Aided Mol.Des., 36, 2022
|
|
7GDN
 
 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with EDJ-MED-0e996074-1 (Mpro-x11159) | | Descriptor: | (4R)-N-(4-cyclopropyl-4H-1,2,4-triazol-3-yl)-3,4-dihydro-2H-1-benzopyran-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5DVC
 
 | | Human PPARgamma ligand binding dmain complexed with SB1453 in a covalent bonded form | | Descriptor: | N-[2-({3-[({4-[(4-methylpiperazin-1-yl)methyl]benzoyl}amino)methyl]benzyl}oxy)phenyl]-3-nitrobenzamide, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Jang, J.Y. | | Deposit date: | 2015-09-21 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Human PPARgamma ligand binding dmain complexed with SB1453 in a covalent bonded form
To Be Published
|
|
