7YRD
 
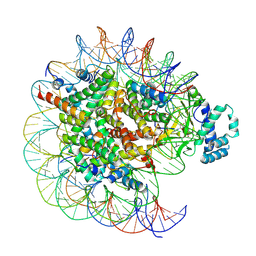 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
7GFJ
 
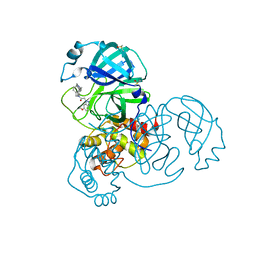 | | Group deposition SARS-CoV-2 main protease in complex with inhibitors from the COVID Moonshot -- Crystal Structure of SARS-CoV-2 main protease in complex with ALP-POS-c0c213c9-1 (Mpro-x11757) | | Descriptor: | 2-(6-methoxy-1H-benzotriazol-1-yl)-N-[4-(piperidin-4-yl)phenyl]-N-[(pyridin-2-yl)methyl]acetamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | Authors: | Fearon, D, Aimon, A, Aschenbrenner, J.C, Balcomb, B.H, Bertram, F.K.R, Brandao-Neto, J, Dias, A, Douangamath, A, Dunnett, L, Godoy, A.S, Gorrie-Stone, T.J, Koekemoer, L, Krojer, T, Lithgo, R.M, Lukacik, P, Marples, P.G, Mikolajek, H, Nelson, E, Owen, C.D, Powell, A.J, Rangel, V.L, Skyner, R, Strain-Damerell, C.M, Thompson, W, Tomlinson, C.W.E, Wild, C, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-08-11 | | Release date: | 2023-11-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Open science discovery of potent noncovalent SARS-CoV-2 main protease inhibitors.
Science, 382, 2023
|
|
5BMU
 
 | |
7D9U
 
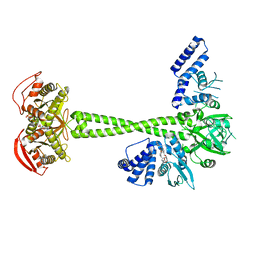 | | Structure of human soluble guanylate cyclase in the cinciguat-bound activated state | | Descriptor: | 4-({(4-carboxybutyl)[2-(2-{[4-(2-phenylethyl)benzyl]oxy}phenyl)ethyl]amino}methyl)benzoic acid, Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1, ... | | Authors: | Chen, L, Liu, R, Kang, Y. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Activation mechanism of human soluble guanylate cyclase by stimulators and activators.
Nat Commun, 12, 2021
|
|
7D9T
 
 | | Structure of human soluble guanylate cyclase in the cinciguat-bound inactive state | | Descriptor: | 4-({(4-carboxybutyl)[2-(2-{[4-(2-phenylethyl)benzyl]oxy}phenyl)ethyl]amino}methyl)benzoic acid, Guanylate cyclase soluble subunit alpha-1, Guanylate cyclase soluble subunit beta-1 | | Authors: | Chen, L, Liu, R, Kang, Y. | | Deposit date: | 2020-10-14 | | Release date: | 2021-08-11 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Activation mechanism of human soluble guanylate cyclase by stimulators and activators.
Nat Commun, 12, 2021
|
|
2Q9A
 
 | | Structure of Apo FTSY | | Descriptor: | 1,2-ETHANEDIOL, Cell division protein ftsY, SULFATE ION | | Authors: | Reyes, C.L, Stroud, R.M. | | Deposit date: | 2007-06-12 | | Release date: | 2007-07-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | X-ray Structures of the Signal Recognition Particle Receptor Reveal Targeting Cycle Intermediates.
Plos One, 2, 2007
|
|
1MNE
 
 | |
2PZA
 
 | | NAD+ Synthetase from Bacillus anthracis with AMP + PPi and Mg2+ | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | McDonald, H.M, Pruett, P.S, Deivanayagam, C, Protasevich, I.I, Carson, W.M, DeLucas, L.J, Brouillette, W.J, Brouillette, C.G. | | Deposit date: | 2007-05-17 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural adaptation of an interacting non-native C-terminal helical extension revealed in the crystal structure of NAD(+) synthetase from Bacillus anthracis.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3JQM
 
 | | Binding of 5'-GTP to molybdenum cofactor biosynthesis protein MoaC from Thermus theromophilus HB8 | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanaujia, S.P, Jeyakanthan, J, Nakagawa, N, Sekar, K, Shinkai, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-09-07 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of apo and GTP-bound molybdenum cofactor biosynthesis protein MoaC from Thermus thermophilus HB8
Acta Crystallogr.,Sect.D, 66, 2010
|
|
5EJV
 
 | | RORy in complex with T090131718 and Coactivator peptide EBI96 | | Descriptor: | EBI96 Coactivator Peptide, N-(2,2,2-TRIFLUOROETHYL)-N-{4-[2,2,2-TRIFLUORO-1-HYDROXY-1-(TRIFLUOROMETHYL)ETHYL]PHENYL}BENZENESULFONAMIDE, Nuclear receptor ROR-gamma | | Authors: | Marcotte, D.M. | | Deposit date: | 2015-11-02 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Discovery of biaryls as ROR gamma inverse agonists by using structure-based design.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
7YRG
 
 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-12-13 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
5ZG1
 
 | | Crystal structure of the GluA2o LBD in complex with glutamate and Compound-2 | | Descriptor: | 9-(4-~{tert}-butylphenyl)-3,4-dihydropyrido[2,1-c][1,2,4]thiadiazine 2,2-dioxide, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2018-03-07 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | TAK-137, an AMPA-R potentiator with little agonistic effect, has a wide therapeutic window.
Neuropsychopharmacology, 44, 2019
|
|
6V0P
 
 | | PRMT5 complex bound to covalent PBM inhibitor BRD6711 | | Descriptor: | 2-(5-chloro-6-oxopyridazin-1(6H)-yl)-N-(4-methyl-3-sulfamoylphenyl)acetamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | McMillan, B.J, McKinney, D.C. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery of a First-in-Class Inhibitor of the PRMT5-Substrate Adaptor Interaction.
J.Med.Chem., 64, 2021
|
|
6VDN
 
 | | HCV NS3/4A protease A156T mutant | | Descriptor: | GLYCEROL, Non-structural protein 4A, Serine protease NS3, ... | | Authors: | Timm, J, Schiffer, C.A. | | Deposit date: | 2019-12-27 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | apo HCV protease A156T
To Be Published
|
|
7ZT2
 
 | | Structure of E8 TCR in complex with human MR1 bound to 5-OP-RU | | Descriptor: | 1,2-ETHANEDIOL, 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, ... | | Authors: | Karuppiah, V, Srikannathasan, V, Robinson, R.A. | | Deposit date: | 2022-05-09 | | Release date: | 2023-06-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Promiscuous recognition of MR1 drives self-reactive mucosal-associated invariant T cell responses.
J.Exp.Med., 220, 2023
|
|
4MC8
 
 | | Hedycaryol synthase in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Putative sesquiterpene cyclase | | Authors: | Baer, P, Rabe, P, Cirton, C, Oliveira Mann, C, Kaufmann, N, Groll, M, Dickschat, J. | | Deposit date: | 2013-08-21 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Hedycaryol synthase in complex with nerolidol reveals terpene cyclase mechanism.
Chembiochem, 15, 2014
|
|
7FJY
 
 | | PanDDA analysis group deposition -- Aar2/RNaseH in complex with fragment P04A02 from the F2X-Universal Library | | Descriptor: | 1-[(3-aminophenyl)methyl]piperidin-4-ol, A1 cistron-splicing factor AAR2, Pre-mRNA-splicing factor 8 | | Authors: | Barthel, T, Wollenhaupt, J, Lima, G.M.A, Wahl, M.C, Weiss, M.S. | | Deposit date: | 2022-08-26 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Large-Scale Crystallographic Fragment Screening Expedites Compound Optimization and Identifies Putative Protein-Protein Interaction Sites.
J.Med.Chem., 65, 2022
|
|
7M0E
 
 | | Pre-catalytic synaptic complex of DNA Polymerase Lambda with gapped DSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DNA (5'-D(*CP*AP*GP*TP*GP*C)-3'), ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
5VM7
 
 | | Pseudo-atomic model of the MurA-A2 complex | | Descriptor: | Maturation protein A2, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Cui, Z, Zhang, J. | | Deposit date: | 2017-04-26 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structures of Q beta virions, virus-like particles, and the Q beta-MurA complex reveal internal coat proteins and the mechanism of host lysis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3GCZ
 
 | | Yokose virus Methyltransferase in complex with AdoMet | | Descriptor: | GLYCEROL, Polyprotein, S-ADENOSYLMETHIONINE, ... | | Authors: | Bollati, M, Milani, M, Mastrangelo, E, Bolognesi, M, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2009-02-23 | | Release date: | 2009-03-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a methyltransferase from a no-known-vector Flavivirus
Biochem.Biophys.Res.Commun., 382, 2009
|
|
7ZNX
 
 | |
6CMS
 
 | | Closed structure of active SHP2 mutant E76K bound to SHP099 inhibitor | | Descriptor: | 6-(4-azanyl-4-methyl-piperidin-1-yl)-3-[2,3-bis(chloranyl)phenyl]pyrazin-2-amine, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Padua, R.A.P, Sun, Y, Marko, I, Pitsawong, W, Kern, D. | | Deposit date: | 2018-03-06 | | Release date: | 2018-11-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Mechanism of activating mutations and allosteric drug inhibition of the phosphatase SHP2.
Nat Commun, 9, 2018
|
|
5I9I
 
 | | Crystal structure of LP_PLA2 in complex with Darapladib | | Descriptor: | N-[2-(diethylamino)ethyl]-2-{2-[(4-fluorobenzyl)sulfanyl]-4-oxo-4,5,6,7-tetrahydro-1H-cyclopenta[d]pyrimidin-1-yl}-N-{[ 4'-(trifluoromethyl)biphenyl-4-yl]methyl}acetamide, Platelet-activating factor acetylhydrolase, SULFATE ION | | Authors: | Liu, Q.F, Xu, Y.C. | | Deposit date: | 2016-02-20 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Thermodynamic Characterization of Protein-Ligand Interactions Formed between Lipoprotein-Associated Phospholipase A2 and Inhibitors
J.Med.Chem., 59, 2016
|
|
2XPK
 
 | | Cell-penetrant, nanomolar O-GlcNAcase inhibitors selective against lysosomal hexosaminidases | | Descriptor: | N-[(5R,6R,7R,8S)-6,7-DIHYDROXY-5-(HYDROXYMETHYL)-2-(2-PHENYLETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDIN-8-YL]-3-SULFANYLPROPANAMIDE, O-GLCNACASE NAGJ | | Authors: | Dorfmueller, H.C, Borodkin, V.S, Schimpl, M, Zheng, X, Kime, R, Read, K.D, van Aalten, D.M.F. | | Deposit date: | 2010-08-26 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cell-Penetrant, Nanomolar O-Glcnacase Inhibitors Selective Against Lysosomal Hexosaminidases.
Chem.Biol, 17, 2010
|
|
5VIX
 
 | |
