6DDR
 
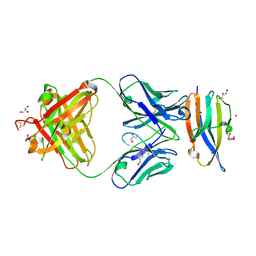 | | Crystal Structure Analysis of the Epitope of an Anti-MICA Antibody | | Descriptor: | Anti-MICA Fab fragment heavy chain clone 13A9, Anti-MICA Fab fragment light chain clone 13A9, GLYCEROL, ... | | Authors: | Matsumoto, M.L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-10-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-resolution glycosylation site-engineering method identifies MICA epitope critical for shedding inhibition activity of anti-MICA antibodies.
MAbs, 11, 2019
|
|
6DDM
 
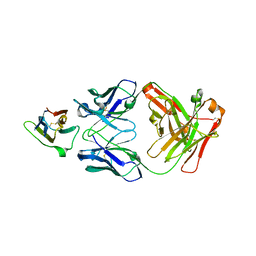 | | Crystal Structure Analysis of the Epitope of an Anti-MICA Antibody | | Descriptor: | Anti-MICA Fab fragment heavy chain clone 1D5, Anti-MICA Fab fragment light chain clone 1D5, MHC class I polypeptide-related sequence A | | Authors: | Matsumoto, M.L. | | Deposit date: | 2018-05-10 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution glycosylation site-engineering method identifies MICA epitope critical for shedding inhibition activity of anti-MICA antibodies.
MAbs, 11, 2019
|
|
7ZI4
 
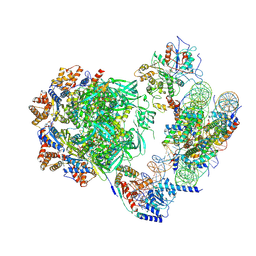 | | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Vance, N.R, Ayala, R, Willhoft, O, Tvardovskiy, A, McCormack, E.A, Bartke, T, Zhang, X, Wigley, D.B. | | Deposit date: | 2022-04-07 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the human INO80 complex bound to a WT nucleosome
To Be Published
|
|
6FGQ
 
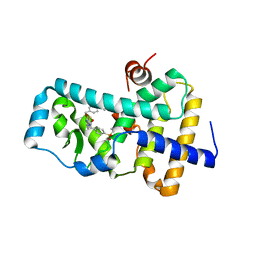 | | Ligand complex of RORg LBD | | Descriptor: | Nuclear receptor ROR-gamma, methyl 4-[[3-[5-[2-(4-ethylsulfonylphenyl)ethanoylamino]thiophen-3-yl]pyridin-2-yl]oxymethyl]benzoate | | Authors: | Xue, Y, Aagaard, A, Narjes, F. | | Deposit date: | 2018-01-11 | | Release date: | 2018-08-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Potent and Orally Bioavailable Inverse Agonists of ROR gamma t Resulting from Structure-Based Design.
J. Med. Chem., 61, 2018
|
|
3G2S
 
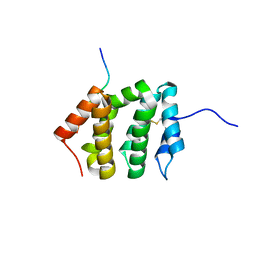 | | VHS Domain of human GGA1 complexed with SorLA C-terminal Peptide | | Descriptor: | ADP-ribosylation factor-binding protein GGA1, C-terminal fragment of Sortilin-related receptor, HEXANE-1,6-DIOL | | Authors: | Cramer, J.F, Behrens, M.A, Gustafsen, C, Oliveira, C.L.P, Pedersen, J.S, Madsen, P, Petersen, C.M, Thirup, S.S. | | Deposit date: | 2009-02-01 | | Release date: | 2009-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | GGA autoinhibition revisited
Traffic, 11, 2010
|
|
6NZD
 
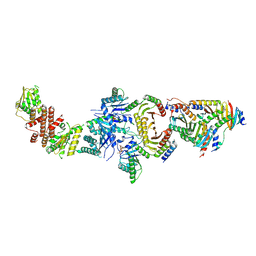 | | Cryo-EM Structure of the Lysosomal Folliculin Complex (FLCN-FNIP2-RagA-RagC-Ragulator) | | Descriptor: | 9-{5-O-[(S)-hydroxy{[(R)-hydroxy(thiophosphonooxy)phosphoryl]oxy}phosphoryl]-alpha-L-arabinofuranosyl}-3,9-dihydro-1H-purine-2,6-dione, Folliculin, Folliculin-interacting protein 2, ... | | Authors: | Fromm, S.A, Young, L.N, Hurley, J.H. | | Deposit date: | 2019-02-13 | | Release date: | 2019-11-06 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural mechanism of a Rag GTPase activation checkpoint by the lysosomal folliculin complex.
Science, 366, 2019
|
|
5J9Q
 
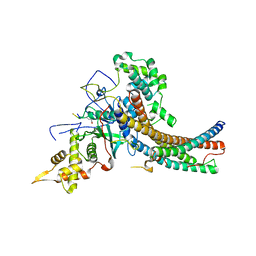 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
6ZM6
 
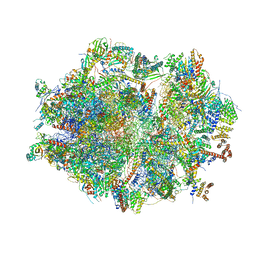 | | Human mitochondrial ribosome in complex with mRNA, A/A tRNA and P/P tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Itoh, Y, Andrell, J, Amunts, A. | | Deposit date: | 2020-07-01 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Mechanism of membrane-tethered mitochondrial protein synthesis.
Science, 371, 2021
|
|
5DE2
 
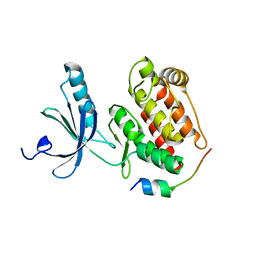 | |
6EHR
 
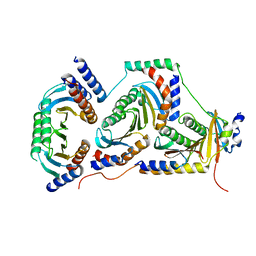 | |
7EQ2
 
 | | Crystal structure of GDP-bound Rab1a-T75D | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CACODYLATE ION, COBALT HEXAMMINE(III), ... | | Authors: | Cao, Y.L, Gu, D.D, Gao, S. | | Deposit date: | 2021-04-28 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55090284 Å) | | Cite: | Aurora kinase A-mediated phosphorylation triggers structural alteration of Rab1A to enhance ER complexity during mitosis
Nat.Struct.Mol.Biol., 2024
|
|
7EU4
 
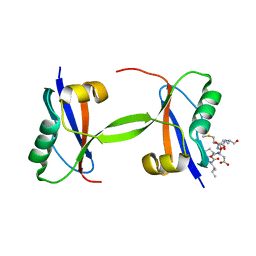 | |
7UX2
 
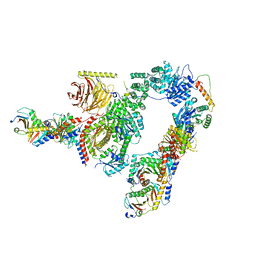 | |
9FMR
 
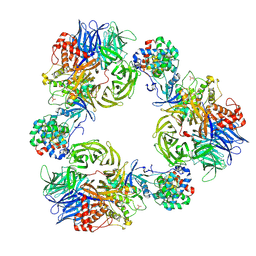 | | Structure of DDB1/Cdk12/Cyclin K with molecular glue SR-4835 | | Descriptor: | Cyclin-K, Cyclin-dependent kinase 12, DNA damage-binding protein 1, ... | | Authors: | Anand, K, Schmitz, M, Geyer, M. | | Deposit date: | 2024-06-07 | | Release date: | 2024-10-16 | | Last modified: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Discovery and design of molecular glue enhancers of CDK12-DDB1 interactions for targeted degradation of cyclin K.
Rsc Chem Biol, 6, 2024
|
|
3SQF
 
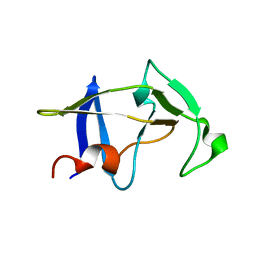 | | Crystal structure of monomeric M-PMV retroviral protease | | Descriptor: | Protease | | Authors: | Jaskolski, M, Kazmierczyk, M, Gilski, M, Krzywda, S, Pichova, I, Zabranska, H, Khatib, F, DiMaio, F, Cooper, S, Thompson, J, Popovic, Z, Baker, D, Group, Foldit Contenders | | Deposit date: | 2011-07-05 | | Release date: | 2011-09-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6324 Å) | | Cite: | Crystal structure of a monomeric retroviral protease solved by protein folding game players.
Nat.Struct.Mol.Biol., 18, 2011
|
|
9B4T
 
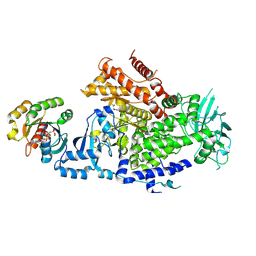 | | Crystal structure of the MRAS-p110alpha complex | | Descriptor: | (2S)-2-({2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}oxy)propanamide, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Czyzyk, D.J, Simanshu, D.K. | | Deposit date: | 2024-03-21 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural insights into isoform-specific RAS-PI3K alpha interactions and the role of RAS in PI3K alpha activation.
Nat Commun, 16, 2025
|
|
7UZO
 
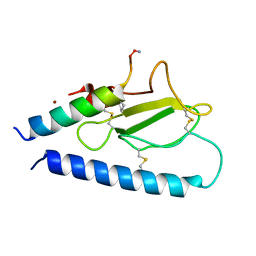 | | Parathyroid hormone 1 receptor extracellular domain complexed with a peptide ligand containing one beta-amino acid | | Descriptor: | Parathyroid hormone/parathyroid hormone-related peptide receptor, Peptide from Parathyroid hormone-related protein, ZINC ION | | Authors: | Yu, Z, Bruchs, A.T, Bingman, C.A, Gellman, S.H. | | Deposit date: | 2022-05-09 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Altered signaling at the PTH receptor via modified agonist contacts with the extracellular domain provides a path to prolonged agonism in vivo.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7UXH
 
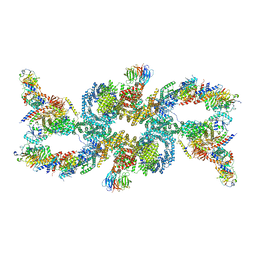 | | cryo-EM structure of the mTORC1-TFEB-Rag-Ragulator complex | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Cui, Z, Hurley, J. | | Deposit date: | 2022-05-05 | | Release date: | 2022-11-30 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the lysosomal mTORC1-TFEB-Rag-Ragulator megacomplex.
Nature, 614, 2023
|
|
7UXC
 
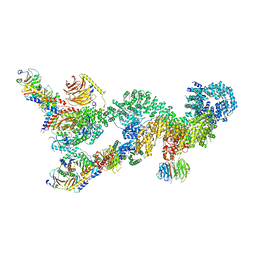 | |
4B9D
 
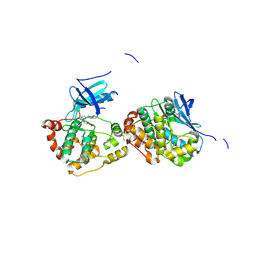 | | Crystal Structure of Human NIMA-related Kinase 1 (NEK1) with inhibitor. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, SERINE/THREONINE-PROTEIN KINASE NEK1, ... | | Authors: | Elkins, J.M, Hanchuk, T.D.M, Lovato, D.V, Basei, F.L, Meirelles, G.V, Kobarg, J, Szklarz, M, Vollmar, M, Mahajan, P, Rellos, P, Zhang, Y, Krojer, T, Pike, A.C.W, Canning, P, von Delft, F, Raynor, J, Bountra, C, Arrowsmith, C, Edwards, A, Knapp, S. | | Deposit date: | 2012-09-04 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | NEK1 kinase domain structure and its dynamic protein interactome after exposure to Cisplatin.
Sci Rep, 7, 2017
|
|
7W98
 
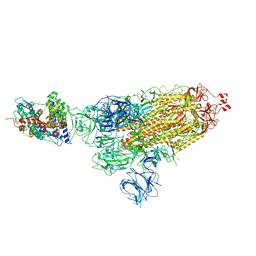 | | SARS-CoV-2 Delta S-ACE2-C1 | | Descriptor: | Angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
7W9I
 
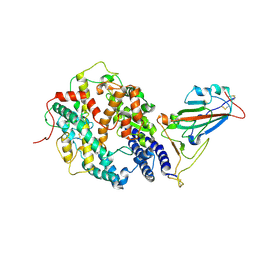 | | SARS-CoV-2 Delta S-RBD-ACE2 | | Descriptor: | Angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Cong, Y, Liu, C.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for SARS-CoV-2 Delta variant recognition of ACE2 receptor and broadly neutralizing antibodies.
Nat Commun, 13, 2022
|
|
8R5J
 
 | | Crystal structure of MERS-CoV main protease | | Descriptor: | Non-structural protein 11 | | Authors: | Balcomb, B.H, Fairhead, M, Koekemoer, L, Lithgo, R.M, Aschenbrenner, J.C, Chandran, A.V, Godoy, A.S, Lukacik, P, Marples, P.G, Mazzorana, M, Ni, X, Strain-Damerell, C, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-11-16 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of MERS-CoV main protease
To Be Published
|
|
4UAL
 
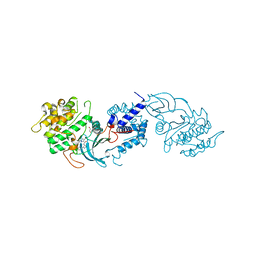 | | MRCK beta in complex with BDP00005290 | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-1-(piperidin-4-yl)-N-[3-(pyridin-2-yl)-1H-pyrazol-4-yl]-1H-pyrazole-3-carboxamide, CHLORIDE ION, ... | | Authors: | Schuettelkopf, A.W. | | Deposit date: | 2014-08-10 | | Release date: | 2014-10-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | A novel small-molecule MRCK inhibitor blocks cancer cell invasion.
Cell Commun. Signal, 12, 2014
|
|
7VVU
 
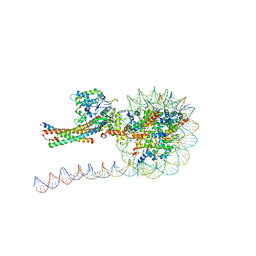 | | NuA4 HAT module bound to the nucleosome | | Descriptor: | CARBOXYMETHYL COENZYME *A, Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, ... | | Authors: | Chen, Z, Qu, K. | | Deposit date: | 2021-11-09 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the NuA4 acetyltransferase complex bound to the nucleosome.
Nature, 610, 2022
|
|
