3TPP
 
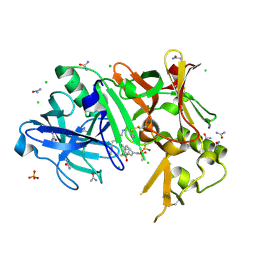 | | Crystal structure of BACE1 complexed with an inhibitor | | Descriptor: | Beta-secretase 1, CHLORIDE ION, N-[(1S,2R)-1-BENZYL-3-(CYCLOPROPYLAMINO)-2-HYDROXYPROPYL]-5-[METHYL(METHYLSULFONYL)AMINO]-N'-[(1R)-1-PHENYLETHYL]ISOPHTHALAMIDE, ... | | Authors: | Xu, Y.C, Li, M.J, Greenblatt, H, Chen, T.T, Silman, I, Sussman, J.L. | | Deposit date: | 2011-09-08 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Flexibility of the flap in the active site of BACE1 as revealed by crystal structures and molecular dynamics simulations
Acta Crystallogr.,Sect.D, 68, 2012
|
|
6T5X
 
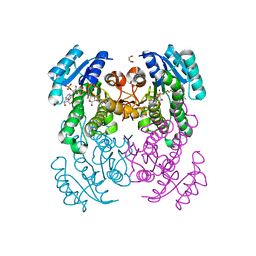 | | Crystal structure of Salmonella typhimurium FabG in complex with NADPH at 1.5 A resolution | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Vella, P, Schnell, R, Schneider, G. | | Deposit date: | 2019-10-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
5J76
 
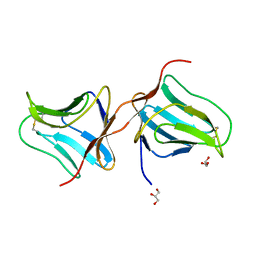 | | Structure of Lectin from Colocasia esculenta(L.) Schott | | Descriptor: | 12kD storage protein, GLYCEROL | | Authors: | Vajravijayan, S, Pletnev, S, Nandhagopal, N, Gunasekaran, K. | | Deposit date: | 2016-04-06 | | Release date: | 2016-06-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural analysis of beta-prism lectin from Colocasia esculenta (L.) S chott.
Int.J.Biol.Macromol., 91, 2016
|
|
6T6P
 
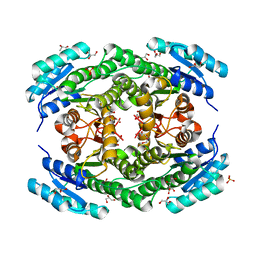 | | Crystal structure of Klebsiella pneumoniae FabG2(NADH-dependent) at 1.57 A resolution | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, GLYCEROL, PHOSPHATE ION | | Authors: | Vella, P, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2019-10-18 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | A FabG inhibitor targeting an allosteric binding site inhibits several orthologs from Gram-negative ESKAPE pathogens.
Bioorg.Med.Chem., 30, 2021
|
|
6T7M
 
 | |
7Q06
 
 | | Crystal structure of TPADO in complex with 2-OH-TPA | | Descriptor: | 2-Hydroxyterephthalic acid, FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7Q05
 
 | | Crystal structure of TPADO in complex with TPA | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Lysozyme, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6NFF
 
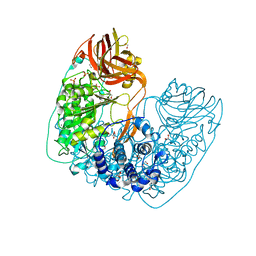 | | Structure of X-prolyl dipeptidyl aminopeptidase from Lactobacillus helveticus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Juers, D.H, Bratt, N.J, Ojennus, D.D. | | Deposit date: | 2018-12-20 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of a prolyl aminodipeptidase (PepX) from Lactobacillus helveticus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
7Q04
 
 | | Crystal structure of TPADO in a substrate-free state | | Descriptor: | FE (III) ION, FE2/S2 (INORGANIC) CLUSTER, Lysozyme, ... | | Authors: | Zahn, M, Kincannon, W.M, DuBois, J.L, McGeehan, J.E. | | Deposit date: | 2021-10-14 | | Release date: | 2022-04-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | Biochemical and structural characterization of an aromatic ring-hydroxylating dioxygenase for terephthalic acid catabolism.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3RLH
 
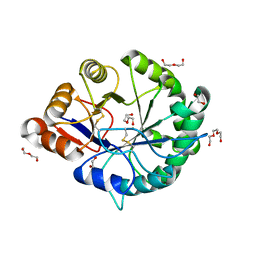 | | Crystal structure of a class II phospholipase D from Loxosceles intermedia venom | | Descriptor: | (2r,5r)-5-amino-2-hydroxy-5-(hydroxymethyl)-1,3,2lambda~5~-dioxaphosphinan-2-one, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Giuseppe, P.O, Ullah, A, Veiga, S.S, Murakami, M.T, Arni, R.K, Doherty, D.Z, Gismene, C, Bachega, J.F.R, Chahine, J, Gonzalez, J.E.H. | | Deposit date: | 2011-04-19 | | Release date: | 2011-06-29 | | Last modified: | 2025-02-19 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure of a novel class II phospholipase D: Catalytic cleft is modified by a disulphide bridge.
Biochem.Biophys.Res.Commun., 409, 2011
|
|
6PT8
 
 | | Crystal Structure of CobT from Methanocaldococcus jannaschii in complex with Adenine Alpha-Ribotide and Nicotinic Acid | | Descriptor: | ALPHA-ADENOSINE MONOPHOSPHATE, NICOTINIC ACID, PHOSPHATE ION, ... | | Authors: | Schwarzwalder, A.H, Jeter, V.L, Vecellio, A.A, Erpenbach, E, Escalante, J.C, Rayment, I. | | Deposit date: | 2019-07-15 | | Release date: | 2020-07-22 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies of the phosphoribosyltransferase involved in cobamide biosynthesis in methanogenic archaea and cyanobacteria.
Sci Rep, 12, 2022
|
|
6TYR
 
 | |
6TV9
 
 | |
8DTA
 
 | |
6UBQ
 
 | |
6TZD
 
 | |
6UCW
 
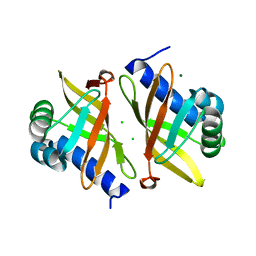 | | Multi-conformer model of Apo Ketosteroid Isomerase from Pseudomonas Putida (pKSI) at 250 K | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Steroid Delta-isomerase | | Authors: | Yabukarski, F, Herschlag, D, Biel, J.T, Fraser, J.S. | | Deposit date: | 2019-09-17 | | Release date: | 2020-09-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Assessment of enzyme active site positioning and tests of catalytic mechanisms through X-ray-derived conformational ensembles.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6U4I
 
 | |
6U1Z
 
 | |
8VSK
 
 | | Crystal structure of Dehaloperoxidase A in complex with substrate 2,4-dibromophenol | | Descriptor: | 2,4-bis(bromanyl)phenol, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-24 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.515 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8VKC
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrophenol | | Descriptor: | Dehaloperoxidase A, GLYCEROL, P-NITROPHENOL, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8VKD
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-nitrocatechol | | Descriptor: | 4-NITROCATECHOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-01-08 | | Release date: | 2024-07-17 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
8VZR
 
 | | Crystal structure of dehaloperoxidase A in complex with substrate 4-bromo-o-cresol | | Descriptor: | 4-bromo-2-methylphenol, DI(HYDROXYETHYL)ETHER, Dehaloperoxidase A, ... | | Authors: | Aktar, M.S, de Serrano, V.S, Ghiladi, R.A, Franzen, S. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Comparison of Substrate Binding Sites in Dehaloperoxidase A and B.
Biochemistry, 63, 2024
|
|
6U60
 
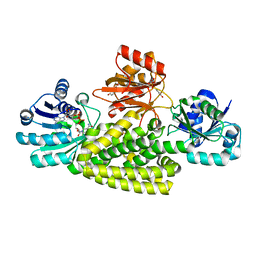 | | Crystal structure of prephenate dehydrogenase tyrA from Bacillus anthracis in complex with NAD and L-tyrosine | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PHOSPHATE ION, Prephenate dehydrogenase, ... | | Authors: | Shabalin, I.G, Hou, J, Kutner, J, Grimshaw, S, Christendat, D, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-28 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical analysis of Bacillus anthracis prephenate dehydrogenase reveals an unusual mode of inhibition by tyrosine via the ACT domain.
Febs J., 287, 2020
|
|
6XR3
 
 | |
