1L35
 
 | |
6FII
 
 | | Tubulin-Spongistatin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Menchon, G, Prota, A.E, Lucena Angell, D, Bucher, P, Mueller, R, Paterson, I, Diaz, J.F, Altmann, K.-H, Steinmetz, M.O. | | Deposit date: | 2018-01-18 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | A fluorescence anisotropy assay to discover and characterize ligands targeting the maytansine site of tubulin.
Nat Commun, 9, 2018
|
|
1L56
 
 | |
4RSS
 
 | | Crystal structure of tyrosine-protein kinase SYK with an inhibitor | | Descriptor: | 1-[(3-methyl-1-{2-[(1,2,3-trimethyl-1H-indol-5-yl)amino]pyrimidin-4-yl}-1H-pyrazol-4-yl)methyl]azetidin-3-ol, Tyrosine-protein kinase SYK | | Authors: | Lee, B.I, Lee, S.J, Choi, J.-S. | | Deposit date: | 2014-11-11 | | Release date: | 2015-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Highly potent and selective pyrazolylpyrimidines as Syk kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7MK0
 
 | |
8YJC
 
 | | Structure of Vibrio vulnificus MARTX cysteine protease domain C3727A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, INOSITOL HEXAKISPHOSPHATE, Multifunctional autoprocessing repeat-in-toxin (MARTX), ... | | Authors: | Chen, L, Khan, H, Tan, L, Li, X, Zhang, G, Im, Y.J. | | Deposit date: | 2024-03-01 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural basis of the activation of MARTX cysteine protease domain from Vibrio vulnificus.
Plos One, 19, 2024
|
|
4C4E
 
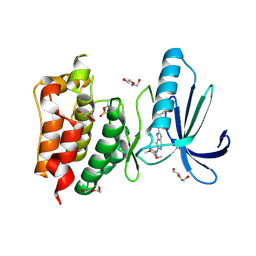 | | Structure-based design of orally bioavailable pyrrolopyridine inhibitors of the mitotic kinase MPS1 | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, DUAL SPECIFICITY PROTEIN KINASE TTK, N-(3,4-dimethoxyphenyl)-2-(1H-pyrazol-4-yl)-1H-pyrrolo[3,2-c]pyridin-6-amine, ... | | Authors: | Naud, S, Westwood, I.M, Faisal, A, Sheldrake, P, Bavetsias, V, Atrash, B, Liu, M, Hayes, A, Schmitt, J, Wood, A, Choi, V, Boxall, K, Mak, G, Gurden, M, Valenti, M, de Haven Brandon, A, Henley, A, Baker, R, McAndrew, C, Matijssen, B, Burke, R, Eccles, S.A, Raynaud, F.I, Linardopoulos, S, van Montfort, R, Blagg, J. | | Deposit date: | 2013-09-05 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of Orally Bioavailable 1H-Pyrrolo[3, 2-C]Pyridine Inhibitors of the Mitotic Kinase Monopolar Spindle 1 (Mps1).
J.Med.Chem., 56, 2013
|
|
8C0F
 
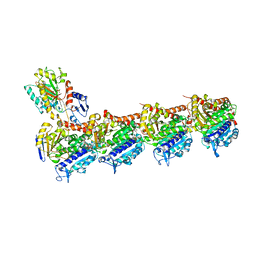 | | Tubulin-PTC596 complex | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5-fluoranyl-2-(6-fluoranyl-2-methyl-benzimidazol-1-yl)-~{N}4-[4-(trifluoromethyl)phenyl]pyrimidine-4,6-diamine, ... | | Authors: | Prota, A.E, Muehlethaler, T, Weetall, M, Steinmetz, M.O. | | Deposit date: | 2022-12-16 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1005 Å) | | Cite: | Preclinical and Early Clinical Development of PTC596, a Novel Small-Molecule Tubulin-Binding Agent
Mol Cancer Ther, 20, 2021
|
|
8CSU
 
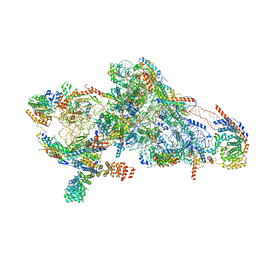 | | Human mitochondrial small subunit assembly intermediate (State C*) | | Descriptor: | 12S mitochondrial rRNA, 28S ribosomal protein S10, mitochondrial, ... | | Authors: | Harper, N.J, Burnside, C, Klinge, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-12-14 | | Last modified: | 2023-02-08 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Principles of mitoribosomal small subunit assembly in eukaryotes.
Nature, 614, 2023
|
|
5HWN
 
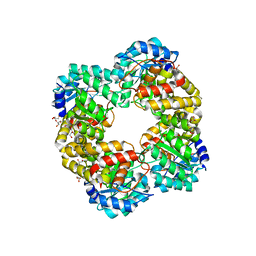 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with pyruvate | | Descriptor: | FORMIC ACID, GLYCEROL, PYRUVIC ACID, ... | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure and function of a decarboxylating Agrobacterium tumefaciens keto-deoxy-d-galactarate dehydratase.
Biochemistry, 53, 2014
|
|
8C01
 
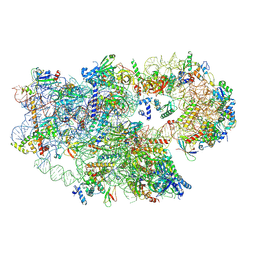 | | Enp1TAP_A population of yeast small ribosomal subunit precursors | | Descriptor: | 18S rRNA precursor, 20S-pre-rRNA D-site endonuclease NOB1, 40S ribosomal protein S0-A, ... | | Authors: | Milkereit, P, Poell, G. | | Deposit date: | 2022-12-15 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Impact of the yeast S0/uS2-cluster ribosomal protein rpS21/eS21 on rRNA folding and the architecture of small ribosomal subunit precursors.
Plos One, 18, 2023
|
|
6TLR
 
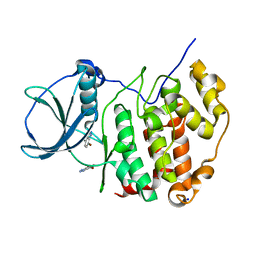 | | HUMAN CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 4,7-DIBROMOBENZOTRIAZOLE | | Descriptor: | 4,7-bis(bromanyl)-1~{H}-benzotriazole, CHLORIDE ION, Casein kinase II subunit alpha, ... | | Authors: | Czapinska, H, Piasecka, A, Winiewska-Szajewska, M, Bochtler, M, Poznanski, J. | | Deposit date: | 2019-12-03 | | Release date: | 2020-12-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Halogen Atoms in the Protein-Ligand System. Structural and Thermodynamic Studies of the Binding of Bromobenzotriazoles by the Catalytic Subunit of Human Protein Kinase CK2.
J.Phys.Chem.B, 125, 2021
|
|
4RW5
 
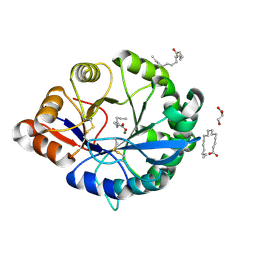 | | Structural insights into substrate binding of brown spider venom class II phospholipases D | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-TRIDECANOIC ACID, ... | | Authors: | Coronado, M.A, Ullah, A, da Silva, L.S, Chaves-Moreira, D, Vuitika, L, Chaim, O.M, Veiga, S.S, Chahine, J, Murakami, M.T, Arni, R.K. | | Deposit date: | 2014-12-01 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural Insights into Substrate Binding of Brown Spider Venom Class II Phospholipases D.
Curr Protein Pept Sci, 16, 2015
|
|
4ZYU
 
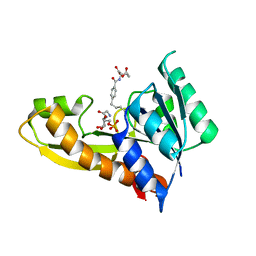 | | Human GAR transformylase in complex with GAR and N-{4-[4-(2-Amino-4-oxo-3,4-dihydrothieno[2,3-d]pyrimidin-6-yl)-butyl]benzoyl}-L-glutamic acid (AGF50) | | Descriptor: | GLYCINAMIDE RIBONUCLEOTIDE, N-{4-[4-(2-amino-4-oxo-1,4-dihydrothieno[2,3-d]pyrimidin-6-yl)butyl]benzoyl}-L-glutamic acid, Trifunctional purine biosynthetic protein adenosine-3 | | Authors: | Deis, S.M, Dann III, C.E. | | Deposit date: | 2015-05-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and Enzymatic Analysis of Tumor-Targeted Antifolates That Inhibit Glycinamide Ribonucleotide Formyltransferase.
Biochemistry, 55, 2016
|
|
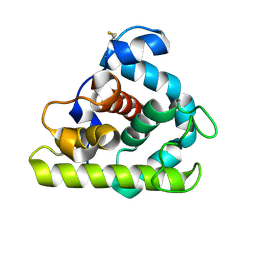
153L
 
 | |
9F2K
 
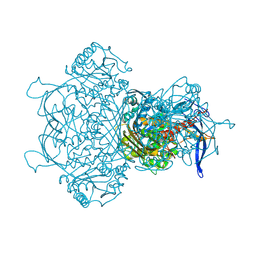 | | Myo-inositol-1-phosphate synthase from Thermochaetoides thermophila in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, inositol-3-phosphate synthase | | Authors: | Traeger, T.K, Kyrilis, F.L, Hamdi, F, Kastritis, P.L. | | Deposit date: | 2024-04-23 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Disorder-to-order active site capping regulates the rate-limiting step of the inositol pathway.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2VRI
 
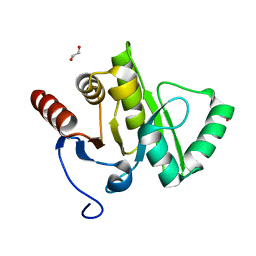 | |
8E43
 
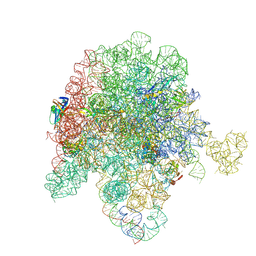 | | E. coli 50S ribosome bound to compound streptogramin A analog 3336 | | Descriptor: | (2R)-2-[(3S,4R,5E,10E,12E,14S,16R,23S,26aR)-16-fluoro-14-hydroxy-12-methyl-1,7,22-trioxo-4-(prop-2-en-1-yl)-4,7,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,3H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosin-3-yl]propyl isoquinolin-3-ylcarbamate, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Seiple, I.B, Fraser, J.S. | | Deposit date: | 2022-08-17 | | Release date: | 2023-06-28 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Streptogramin A analogs
To Be Published
|
|
4CEY
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor NLD | | Descriptor: | 1-(2-aminopyridin-4-yl)-3-[(3S)-5-{4-[(E)-(ethoxyimino)methyl]phenoxy}-3-methylpentyl]imidazolidin-2-one, SODIUM ION, VP1, ... | | Authors: | De Colibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-12 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
6THP
 
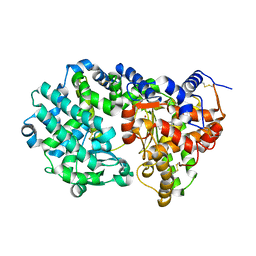 | |
7YAG
 
 | | CryoEM structure of SPCA1a in E1-Ca-AMPPCP state subclass 1 | | Descriptor: | CALCIUM ION, Calcium-transporting ATPase type 2C member 1, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, ... | | Authors: | Chen, Z, Watanabe, S, Inaba, K. | | Deposit date: | 2022-06-28 | | Release date: | 2023-03-22 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human SPCA1a reveal the mechanism of Ca 2+ /Mn 2+ transport into the Golgi apparatus.
Sci Adv, 9, 2023
|
|
8QSJ
 
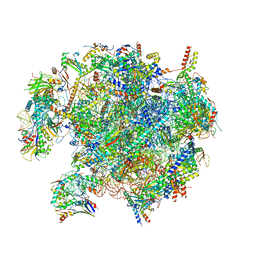 | | Human mitoribosomal large subunit assembly intermediate 2 with GTPBP7 | | Descriptor: | 16S rRNA, 39S ribosomal protein L10, mitochondrial, ... | | Authors: | Ritter, C, Nguyen, T.G, Kummer, E. | | Deposit date: | 2023-10-10 | | Release date: | 2023-12-13 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the role of GTPBP10 in the RNA maturation of the mitoribosome.
Nat Commun, 14, 2023
|
|
7YAJ
 
 | | CryoEM structure of SPCA1a in E1-Mn-AMPPCP state subclass 1 | | Descriptor: | Calcium-transporting ATPase type 2C member 1, MANGANESE (II) ION, Nanobody head piece of megabody, ... | | Authors: | Chen, Z, Watanabe, S, Inaba, K. | | Deposit date: | 2022-06-28 | | Release date: | 2023-03-22 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structures of human SPCA1a reveal the mechanism of Ca 2+ /Mn 2+ transport into the Golgi apparatus.
Sci Adv, 9, 2023
|
|
8T3O
 
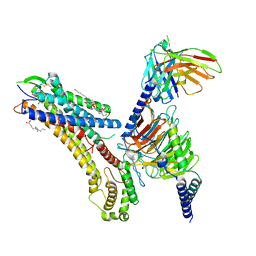 | | Cryo-EM structure of the TUG-891 bound FFA4-Gq complex | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid, Free fatty acid receptor 4, ... | | Authors: | Zhang, X, Tikhonova, I, Milligan, G, Zhang, C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-01-17 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for the ligand recognition and signaling of free fatty acid receptors.
Sci Adv, 10, 2024
|
|
4S18
 
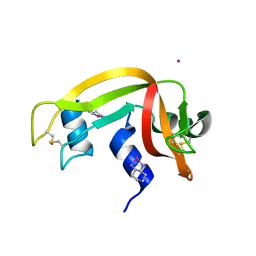 | |
