5WPJ
 
 | |
5M3W
 
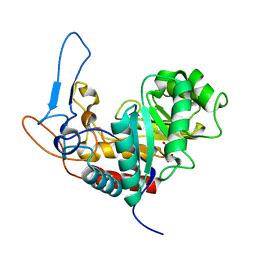 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-1,2-dideoxymannose and alpha-1,2-mannobiose | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-10-17 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
5WQC
 
 | | Crystal structure of human orexin 2 receptor bound to the selective antagonist EMPA determined by the synchrotron light source at SPring-8. | | Descriptor: | N-ethyl-2-[(6-methoxypyridin-3-yl)-(2-methylphenyl)sulfonyl-amino]-N-(pyridin-3-ylmethyl)ethanamide, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Orexin receptor type 2, ... | | Authors: | Suno, R, Hirata, K, Yamashita, K, Tsujimoto, H, Sasanuma, M, Horita, S, Yamamoto, M, Rosenbaum, D.M, Iwata, S, Shimamura, T, Kobayashi, T. | | Deposit date: | 2016-11-25 | | Release date: | 2017-11-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of Human Orexin 2 Receptor Bound to the Subtype-Selective Antagonist EMPA
Structure, 26, 2018
|
|
5LXB
 
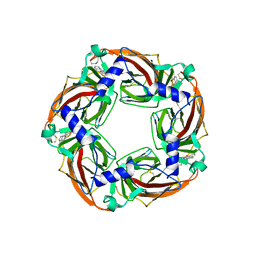 | |
5WRJ
 
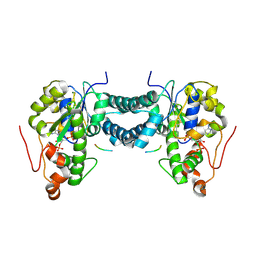 | | Crystal structure of human tyrosylprotein sulfotransferase-1 complexed with PAP and gastrin peptide | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, MAGNESIUM ION, Protein-tyrosine sulfotransferase 1, ... | | Authors: | Tanaka, S, Nishiyori, T, Kojo, H, Otsubo, R, Kakuta, Y. | | Deposit date: | 2016-12-02 | | Release date: | 2017-09-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural basis for the broad substrate specificity of the human tyrosylprotein sulfotransferase-1.
Sci Rep, 7, 2017
|
|
5WS5
 
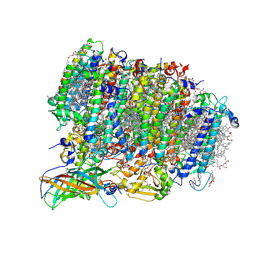 | | Native XFEL structure of photosystem II (preflash dark dataset) | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Shen, J.R. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Light-induced structural changes and the site of O=O bond formation in PSII caught by XFEL.
Nature, 543, 2017
|
|
5X07
 
 | | Crystal structure of FOXA2 DNA binding domain bound to a full consensus DNA site | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Hepatocyte nuclear factor 3-beta | | Authors: | Li, J, Guo, M, Zhou, Z, Jiang, L, Chen, X, Qu, L, Wu, D, Chen, Z, Chen, L, Chen, Y. | | Deposit date: | 2017-01-20 | | Release date: | 2017-08-16 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure of the Forkhead Domain of FOXA2 Bound to a Complete DNA Consensus Site
Biochemistry, 56, 2017
|
|
5X0X
 
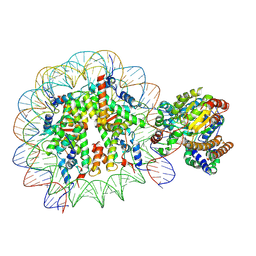 | | Complex of Snf2-Nucleosome complex with Snf2 bound to position +6 of the nucleosome | | Descriptor: | DNA (167-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Li, M, Liu, X, Xia, X, Chen, Z, Li, X. | | Deposit date: | 2017-01-23 | | Release date: | 2017-04-19 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.97 Å) | | Cite: | Mechanism of chromatin remodelling revealed by the Snf2-nucleosome structure.
Nature, 544, 2017
|
|
5WUA
 
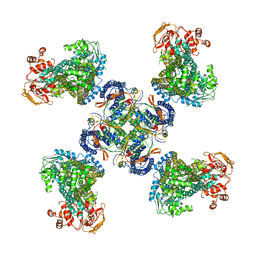 | | Structure of a Pancreatic ATP-sensitive Potassium Channel | | Descriptor: | ATP-sensitive inward rectifier potassium channel 11,superfolder GFP, SUR1 | | Authors: | Li, N, Wu, J.-X, Chen, L, Gao, N. | | Deposit date: | 2016-12-16 | | Release date: | 2017-01-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (5.6 Å) | | Cite: | Structure of a Pancreatic ATP-Sensitive Potassium Channel
Cell, 168, 2017
|
|
5LRB
 
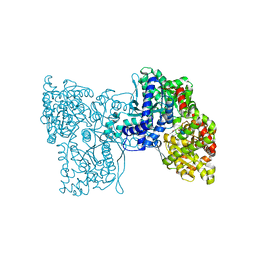 | | Plastidial phosphorylase from Barley in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-1,4 glucan phosphorylase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Cuesta-Seijo, J.A, Ruzanski, C, Kruzewicz, K, Palcic, M.M. | | Deposit date: | 2016-08-18 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional and structural characterization of plastidic starch phosphorylase during barley endosperm development.
PLoS ONE, 12, 2017
|
|
5LK3
 
 | |
6DE7
 
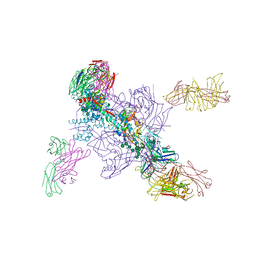 | |
5M36
 
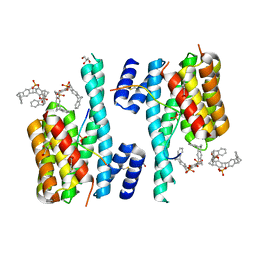 | | The molecular tweezer CLR01 stabilizes a disordered protein-protein interface | | Descriptor: | (1R,5S,9S,16R,20R,24S,28S,35R)-3,22-Bis(dihydroxyphosphoryloxy)tridecacyclo[22.14.1.15,20.19,16.128,35.02,23.04,21.06,19.08,17.010,15.025,38.027,36.029,34]dotetraconta-2(23),3,6,8(17),10,12,14,18,21,25,27(36),29,31,33,37-pentadecaene, 14-3-3 protein zeta/delta, BENZOIC ACID, ... | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2016-10-14 | | Release date: | 2017-11-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Molecular Tweezer CLR01 Stabilizes a Disordered Protein-Protein Interface.
J. Am. Chem. Soc., 139, 2017
|
|
7S3F
 
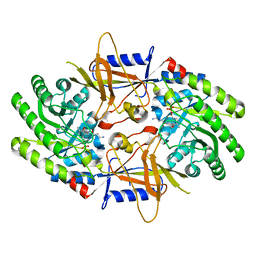 | | Structure of cofactor pyridoxal 5-phosphate bound human ornithine decarboxylase in complex with its inhibitor 1-amino-oxy-3-aminopropane | | Descriptor: | 3-AMINOOXY-1-AMINOPROPANE, Ornithine decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhou, X.E, Suino-Powell, K, Schultz, C.R, Aleiwi, B, Brunzelle, J.S, Lamp, J, Vega, I.E, Ellsworth, E, Bachmann, A.S, Melcher, K. | | Deposit date: | 2021-09-06 | | Release date: | 2021-12-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural basis of binding and inhibition of ornithine decarboxylase by 1-amino-oxy-3-aminopropane.
Biochem.J., 478, 2021
|
|
6D64
 
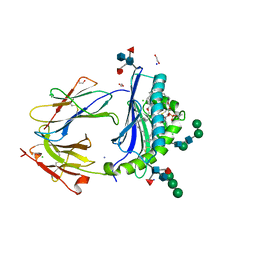 | | Crystal Structure of Human CD1b in Complex with POPC | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 1,2-ETHANEDIOL, Beta-2-microglobulin, ... | | Authors: | Shahine, A.E, Rossjohn, J. | | Deposit date: | 2018-04-20 | | Release date: | 2019-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A T-cell receptor escape channel allows broad T-cell response to CD1b and membrane phospholipids.
Nat Commun, 10, 2019
|
|
7S97
 
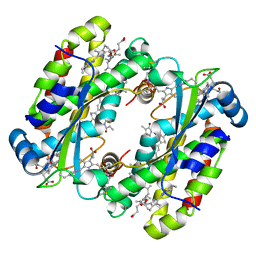 | | Structure of the Photoacclimated Light Harvesting Complex PC577 from Hemiselmis pacifica | | Descriptor: | 15,16-DIHYDROBILIVERDIN, PHYCOCYANOBILIN, Phycoerythrin alpha subunit 1, ... | | Authors: | Jeffrey, P.D, Spangler, L.C, Scholes, G.D. | | Deposit date: | 2021-09-20 | | Release date: | 2022-02-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Controllable Phycobilin Modification: An Alternative Photoacclimation Response in Cryptophyte Algae.
Acs Cent.Sci., 8, 2022
|
|
7SAU
 
 | |
7SHW
 
 | |
5W41
 
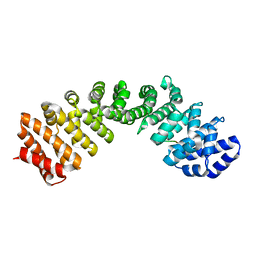 | |
1DQB
 
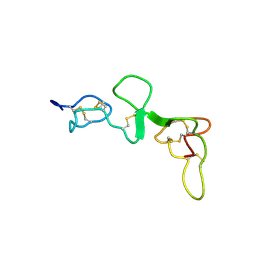 | |
5W5P
 
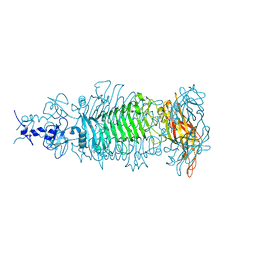 | |
5VVW
 
 | |
5W70
 
 | | X-ray Structure of RbmB from Streptomyces ribosidificus | | Descriptor: | 1,2-ETHANEDIOL, L-glutamine:2-deoxy-scyllo-inosose aminotransferase, [4-({[(1R,2S,3S,4R,5S)-5-amino-2,3,4-trihydroxycyclohexyl]amino}methyl)-5-hydroxy-6-methylpyridin-3-yl]methyl dihydrogen phosphate | | Authors: | Zachman-Brockmeyer, T.R, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-06-19 | | Release date: | 2017-07-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of RbmB from Streptomyces ribosidificus, an aminotransferase involved in the biosynthesis of ribostamycin.
Protein Sci., 26, 2017
|
|
5VWX
 
 | | Bak core latch dimer in complex with Bim-h0-h3Glt | | Descriptor: | 1,2-ETHANEDIOL, Bcl-2 homologous antagonist/killer, Bcl-2-like protein 11 | | Authors: | Brouwer, J.M, Lan, P, Lessene, G, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.489 Å) | | Cite: | Conversion of Bim-BH3 from Activator to Inhibitor of Bak through Structure-Based Design.
Mol. Cell, 68, 2017
|
|
5VX6
 
 | | Structure of Bacillus subtilis Inhibitor of motility (MotI/DgrA) | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Uncharacterized protein YpfA | | Authors: | Subramanian, S, Dann III, C. | | Deposit date: | 2017-05-23 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.197 Å) | | Cite: | MotI (DgrA) acts as a molecular clutch on the flagellar stator protein MotA inBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
