1EDX
 
 | | SOLUTION STRUCTURE OF AMINO TERMINUS OF BOVINE RHODOPSIN (RESIDUES 1-40) | | Descriptor: | RHODOPSIN | | Authors: | Yeagle, P.L, Salloum, A, Chopra, A, Bhawsar, N, Ali, L. | | Deposit date: | 2000-01-28 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the intradiskal loops and amino terminus of the G-protein receptor, rhodopsin.
J.Pept.Res., 55, 2000
|
|
1EDV
 
 | | SOLUTION STRUCTURE OF 2ND INTRADISKAL LOOP OF BOVINE RHODOPSIN (RESIDUES 172-205) | | Descriptor: | RHODOPSIN | | Authors: | Yeagle, P.L, Salloum, A, Chopra, A, Bhawsar, N, Ali, L. | | Deposit date: | 2000-01-28 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the intradiskal loops and amino terminus of the G-protein receptor, rhodopsin.
J.Pept.Res., 55, 2000
|
|
1YUZ
 
 | | Partially Reduced State of Nigerythrin | | Descriptor: | FE (II) ION, FE (III) ION, Nigerythrin | | Authors: | Iyer, R.B, Silaghi-Dumitrescu, R, Kurtz, D.M, Lanzilotta, W.N. | | Deposit date: | 2005-02-14 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution crystal structures of Desulfovibrio vulgaris (Hildenborough) nigerythrin: facile, redox-dependent iron movement, domain interface variability, and peroxidase activity in the rubrerythrins.
J.Biol.Inorg.Chem., 10, 2005
|
|
1TO1
 
 | | crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 Y61A mutant | | Descriptor: | CALCIUM ION, CITRIC ACID, PENTAETHYLENE GLYCOL, ... | | Authors: | Radisky, E.S, Kwan, G, Karen Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Binding, Proteolytic, and Crystallographic Analyses of Mutations at the Protease-Inhibitor Interface of the Subtilisin BPN'/Chymotrypsin Inhibitor 2 Complex(,).
Biochemistry, 43, 2004
|
|
1TO2
 
 | | crystal structure of the complex of subtilisin BPN' with chymotrypsin inhibitor 2 M59K, in pH 9 cryosoak | | Descriptor: | CALCIUM ION, CITRIC ACID, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Radisky, E.S, Kwan, G, Karen Lu, C.J, Koshland Jr, D.E. | | Deposit date: | 2004-06-11 | | Release date: | 2004-11-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Binding, Proteolytic, and Crystallographic Analyses of Mutations at the Protease-Inhibitor Interface of the Subtilisin BPN'/Chymotrypsin Inhibitor 2 Complex(,).
Biochemistry, 43, 2004
|
|
1YV1
 
 | | Fully reduced state of nigerythrin (all ferrous) | | Descriptor: | FE (II) ION, Nigerythrin | | Authors: | Iyer, R.B, Silaghi-Dumitrescu, R, Kurtz, D.M, Lanzilotta, W.N. | | Deposit date: | 2005-02-14 | | Release date: | 2005-06-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High-resolution crystal structures of Desulfovibrio vulgaris (Hildenborough) nigerythrin: facile, redox-dependent iron movement, domain interface variability, and peroxidase activity in the rubrerythrins.
J.Biol.Inorg.Chem., 10, 2005
|
|
2FLU
 
 | | Crystal Structure of the Kelch-Neh2 Complex | | Descriptor: | Kelch-like ECH-associated protein 1, Nrf2 | | Authors: | Li, X, Lo, J, Beamer, L, Hannink, M. | | Deposit date: | 2006-01-06 | | Release date: | 2006-08-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling.
Embo J., 25, 2006
|
|
5M5Q
 
 | | COPS5(2-257) IN COMPLEX WITH A AZAINDOLE (COMPOUND 4) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-[(3~{R})-3-(1~{H}-benzimidazol-2-yl)morpholin-4-yl]-3-[2-(4-methyl-2-phenyl-phenyl)-1~{H}-pyrrolo[2,3-b]pyridin-3-yl]propan-1-one, COP9 signalosome complex subunit 5, ... | | Authors: | Renatus, M, Altmann, E. | | Deposit date: | 2016-10-22 | | Release date: | 2017-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Azaindoles as Zinc-Binding Small-Molecule Inhibitors of the JAMM Protease CSN5.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
2HVA
 
 | | Solution Structure of the haem-binding protein p22HBP | | Descriptor: | Heme-binding protein 1 | | Authors: | Gell, D.A, Mackay, J.P, Westman, B.J, Liew, C.K, Gorman, D. | | Deposit date: | 2006-07-28 | | Release date: | 2006-08-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Novel Haem-binding Interface in the 22 kDa Haem-binding Protein p22HBP.
J.Mol.Biol., 362, 2006
|
|
5NR8
 
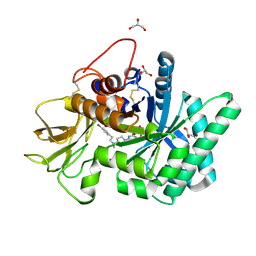 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 7a | | Descriptor: | 1-(3-azanyl-1~{H}-1,2,4-triazol-5-yl)-~{N}-[2-(4-bromophenyl)ethyl]-~{N}-methyl-piperidin-4-amine, Chitotriosidase-1, GLYCEROL | | Authors: | Podjarny, A, Fadel, F, Golebiowski, A. | | Deposit date: | 2017-04-22 | | Release date: | 2018-02-21 | | Last modified: | 2019-10-16 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | Targeting Acidic Mammalian chitinase Is Effective in Animal Model of Asthma.
J. Med. Chem., 61, 2018
|
|
1EEG
 
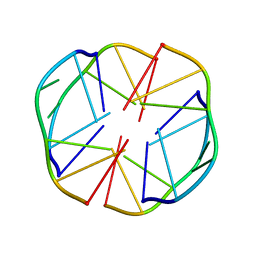 | | A(GGGG)A HEXAD PAIRING ALIGMENT FOR THE D(G-G-A-G-G-A-G) SEQUENCE | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*A)-3') | | Authors: | Kettani, A, Gorin, A, Majumdar, A, Hermann, T, Skripkin, E, Zhao, H, Jones, R, Patel, D.J. | | Deposit date: | 2000-01-31 | | Release date: | 2000-04-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A dimeric DNA interface stabilized by stacked A.(G.G.G.G).A hexads and coordinated monovalent cations.
J.Mol.Biol., 297, 2000
|
|
5NMU
 
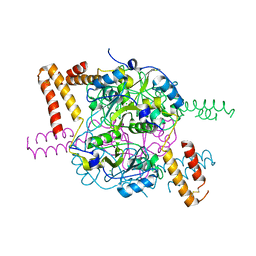 | | Structure of hexameric CBS-CP12 protein from bloom-forming cyanobacteria | | Descriptor: | CBS-CP12, CHLORIDE ION | | Authors: | Hackenberg, C, Hakanpaa, J, Antonyuk, S.V, Dittmann, E, Lamzin, V.S. | | Deposit date: | 2017-04-07 | | Release date: | 2018-05-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2F9J
 
 | |
2C5E
 
 | | gdp-mannose-3', 5' -epimerase (arabidopsis thaliana), k217a, with gdp-alpha-d-mannose bound in the active site. | | Descriptor: | FORMIC ACID, GDP-MANNOSE-3', 5'-EPIMERASE, ... | | Authors: | Major, L.L, Wolucka, B.A, Naismith, J.H. | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Function of Gdp-Mannose-3',5'-Epimerase: An Enzyme which Performs Three Chemical Reactions at the Same Active Site.
J.Am.Chem.Soc., 127, 2005
|
|
5NVD
 
 | | Crystal structure of hexameric CBS-CP12 protein from bloom-forming cyanobacteria at 2.5 A resolution in P6322 crystal form | | Descriptor: | CBS-CP12 | | Authors: | Hackenberg, C, Hakanpaa, J, Eigner, C, Antonyuk, S.V, Dittmann, E, Lamzin, V.S. | | Deposit date: | 2017-05-04 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
2C5A
 
 | | GDP-mannose-3', 5' -epimerase (Arabidopsis thaliana),Y174F, with GDP-beta-L-galactose bound in the active site | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, GDP-MANNOSE-3', ... | | Authors: | Major, L.L, Wolucka, B.A, Naismith, J.H. | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and Function of Gdp-Mannose-3',5'-Epimerase: An Enzyme which Performs Three Chemical Reactions at the Same Active Site.
J.Am.Chem.Soc., 127, 2005
|
|
2LW8
 
 | |
1NMU
 
 | | MBP-L30 | | Descriptor: | 60S ribosomal protein L30, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding periplasmic protein | | Authors: | Chao, J.A, Prasad, G.S, White, S.A, Stout, C.D, Williamson, J.R. | | Deposit date: | 2003-01-10 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Inherent Protein Structural Flexibility at the RNA-binding Interface of L30e
J.Mol.Biol., 326, 2003
|
|
2BTG
 
 | |
2BTH
 
 | |
2C54
 
 | | gdp-mannose-3', 5' -epimerase (arabidopsis thaliana),k178r, with gdp-beta-l-gulose and gdp-4-keto-beta-l-gulose bound in active site. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, GDP-MANNOSE-3', ... | | Authors: | Major, L.L, Wolucka, B.A, Naismith, J.H. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of GDP-mannose-3',5'-epimerase: an enzyme which performs three chemical reactions at the same active site.
J. Am. Chem. Soc., 127, 2005
|
|
2C59
 
 | | gdp-mannose-3', 5' -epimerase (arabidopsis thaliana), with gdp-alpha-d-mannose and gdp-beta-l-galactose bound in the active site. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, GDP-MANNOSE-3', ... | | Authors: | Major, L.L, Wolucka, B.A, Naismith, J.H. | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of Gdp-Mannose-3',5'-Epimerase: An Enzyme which Performs Three Chemical Reactions at the Same Active Site.
J.Am.Chem.Soc., 127, 2005
|
|
1JCA
 
 | | Non-standard Design of Unstable Insulin Analogues with Enhanced Activity | | Descriptor: | ZINC ION, insulin a, insulin b | | Authors: | Weiss, M.A, Wan, Z, Zhao, M, Chu, Y.-C, Nakagawa, S.H, Burke, G.T, Jia, W, Hellmich, R, Katsoyannis, P.G. | | Deposit date: | 2001-06-08 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Non-standard insulin design: structure-activity relationships at the periphery of the insulin receptor.
J.Mol.Biol., 315, 2002
|
|
1P5R
 
 | | Formyl-CoA Transferase in complex with Coenzyme A | | Descriptor: | COENZYME A, Formyl-coenzyme A transferase | | Authors: | Ricagno, S, Jonsson, S, Richards, N, Lindqvist, Y. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Formyl-CoA Transferase encloses the CoA binding site at the interface of an interlocked dimer
Embo J., 22, 2003
|
|
5NPL
 
 | | Crystal structure of hexameric CBS-CP12 protein from bloom-forming cyanobacteria, Yb-derivative at 2.8 A resolution | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, Similar to tr|Q8YYT1|Q8YYT1, YTTERBIUM (III) ION | | Authors: | Hackenberg, C, Hakanpaa, J, Antonyuk, S.V, Dittmann, E, Lamzin, V.S. | | Deposit date: | 2017-04-17 | | Release date: | 2018-05-30 | | Last modified: | 2018-07-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural and functional insights into the unique CBS-CP12 fusion protein family in cyanobacteria.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
