4LWW
 
 | | Discovery of Potent and Efficacious Cyanoguanidine-containing Nicotinamide Phosphoribosyltransferase (Nampt) Inhibitors | | Descriptor: | 1,2-ETHANEDIOL, N-(4-(phenylsulfonyl)benzyl)-1H-pyrrolo[3,2-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Zheng, X, Baumeister, T, Buckmelter, A.J, Caligiuri, M, Clodfelter, K.H, Han, B, Ho, Y, Kley, N, Lin, J, Reynoids, D.J, Sharma, G, Smith, C.C, Wang, Z, Dragovich, P.S, Oh, A, Wang, W, Zak, M, Wang, L, Yuen, P, Bair, K.W. | | Deposit date: | 2013-07-28 | | Release date: | 2013-12-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Discovery of potent and efficacious cyanoguanidine-containing nicotinamide phosphoribosyltransferase (Nampt) inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
7M0D
 
 | | Pre-catalytic quaternary complex of DNA Polymerase Lambda with bound complementary DSB substrate and incoming dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Kaminski, A.M, Bebenek, K, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Analysis of diverse double-strand break synapsis with Pol lambda reveals basis for unique substrate specificity in nonhomologous end-joining.
Nat Commun, 13, 2022
|
|
2XNS
 
 | | Crystal Structure Of Human G alpha i1 Bound To A Designed Helical Peptide Derived From The Goloco Motif Of RGS14 | | Descriptor: | GUANINE NUCLEOTIDE-BINDING PROTEIN G(I) SUBUNIT ALPHA-1, GUANOSINE-5'-DIPHOSPHATE, REGULATOR OF G-PROTEIN SIGNALING 14, ... | | Authors: | Bosch, D, Sammond, D.W, Butterfoss, G.L, Machius, M, Siderovski, D.P, Kuhlman, B. | | Deposit date: | 2010-08-05 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Computational Design of the Sequence and Structure of a Protein-Binding Peptide.
J.Am.Chem.Soc., 133, 2011
|
|
2C5D
 
 | | Structure of a minimal Gas6-Axl complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GROWTH-ARREST-SPECIFIC PROTEIN 6 PRECURSOR, ... | | Authors: | Sasaki, T, Knyazev, P.G, Clout, N.J, Cheburkin, Y, Goehring, W, Ullrich, A, Timpl, R, Hohenester, E. | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis for Gas6-Axl Signalling.
Embo J., 25, 2006
|
|
6VVT
 
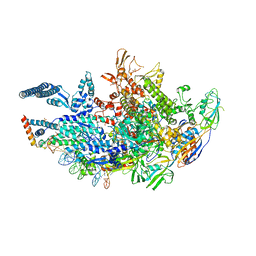 | | Crystal structure of a Mycobacterium smegmatis transcription initiation complex with Rifampicin-resistant RNA polymerase and antibiotic Sorangicin | | Descriptor: | 1,2-ETHANEDIOL, DNA (26-MER), DNA (31-MER), ... | | Authors: | Lilic, M, Darst, S.A, Campbell, E.A. | | Deposit date: | 2020-02-18 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | The antibiotic sorangicin A inhibits promoter DNA unwinding in a Mycobacterium tuberculosis rifampicin-resistant RNA polymerase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
4LXU
 
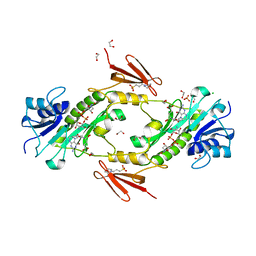 | | dTdp-Fuc3N and 5-N-Formyl-THF | | Descriptor: | (3R,4S,5R,6R)-4-amino-3,5-dihydroxy-6-methyloxan-2-yl][hydroxy-[[(2R,3S,5R)-3-hydroxy-5-(5-methyl-2,4-dioxopyrimidin-1-yl)oxolan-2-yl]methoxy]phosphoryl] hydrogen phosphate, 1,2-ETHANEDIOL, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, ... | | Authors: | Thoden, J.B, Goneau, M.-F, Gilbert, M, Holden, H.M. | | Deposit date: | 2013-07-30 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of a sugar N-formyltransferase from Campylobacter jejuni.
Biochemistry, 52, 2013
|
|
4DVI
 
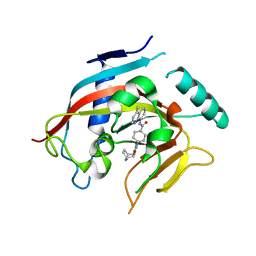 | | Crystal structure of Tankyrase 1 with IWR2 | | Descriptor: | 4-[(3aR,4S,7R,7aS)-1,3-dioxo-1,3,3a,4,7,7a-hexahydro-2H-4,7-methanoisoindol-2-yl]-N-(4-methylquinolin-8-yl)benzamide, Tankyrase-1, ZINC ION | | Authors: | Huang, X. | | Deposit date: | 2012-02-23 | | Release date: | 2012-04-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel binding mode of a potent and selective tankyrase inhibitor.
Plos One, 7, 2012
|
|
7A3H
 
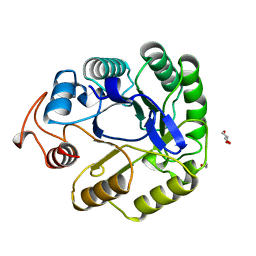 | | NATIVE ENDOGLUCANASE CEL5A CATALYTIC CORE DOMAIN AT 0.95 ANGSTROMS RESOLUTION | | Descriptor: | ENDOGLUCANASE, ETHANOL, GLYCEROL | | Authors: | Davies, G.J, Varrot, A, Dauter, M, Brzozowski, A.M, Schulein, M, Mackenzie, L, Withers, S.G. | | Deposit date: | 1998-08-05 | | Release date: | 1999-08-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Snapshots along an enzymatic reaction coordinate: analysis of a retaining beta-glycoside hydrolase.
Biochemistry, 37, 1998
|
|
1SMD
 
 | | HUMAN SALIVARY AMYLASE | | Descriptor: | AMYLASE, CALCIUM ION, CHLORIDE ION | | Authors: | Ramasubbu, N. | | Deposit date: | 1996-01-24 | | Release date: | 1996-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of human salivary alpha-amylase at 1.6 A resolution: implications for its role in the oral cavity.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
4MA8
 
 | | Crystal structure of mouse prion protein complexed with Chlorpromazine | | Descriptor: | 3-(2-chloro-10H-phenothiazin-10-yl)-N,N-dimethylpropan-1-amine, Major prion protein, POM1 heavy chain, ... | | Authors: | Baral, P.K, Swayampakula, M, James, M.N.G. | | Deposit date: | 2013-08-15 | | Release date: | 2014-01-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of prion inhibition by phenothiazine compounds.
Structure, 22, 2014
|
|
6VU2
 
 | | M1214_N1 Fab structure | | Descriptor: | M1214 N1 Fab heavy chain, M1214 N1 Fab light chain | | Authors: | Pan, R, Kong, X. | | Deposit date: | 2020-02-14 | | Release date: | 2020-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | VSV-Displayed HIV-1 Envelope Identifies Broadly Neutralizing Antibodies Class-Switched to IgG and IgA.
Cell Host Microbe, 27, 2020
|
|
7B6J
 
 | | Crystal structure of MurE from E.coli in complex with minifrag succinimide | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase, ... | | Authors: | Koekemoer, L, Steindel, M, Fairhead, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Krojer, T, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of MurE from E.coli
To Be Published
|
|
4MF1
 
 | | ITK kinase domain in complex with benzothiazole inhibitor 12b (1S,2S)-2-{4-[(DIMETHYLAMINO)METHYL]PHENYL}-N-[6-(1H-PYRAZOL-4-YL)-1,3-BENZOTHIAZOL-2-YL]CYCLOPROPANECARBOXAMIDE | | Descriptor: | (1S,2S)-2-{4-[(dimethylamino)methyl]phenyl}-N-[6-(1H-pyrazol-4-yl)-1,3-benzothiazol-2-yl]cyclopropanecarboxamide, Tyrosine-protein kinase ITK/TSK | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2013-08-27 | | Release date: | 2013-11-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | Structure-based design and synthesis of potent benzothiazole inhibitors of interleukin-2 inducible T cell kinase (ITK).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3ST0
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: halide-free | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-08 | | Release date: | 2012-07-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
3GK7
 
 | | Crystal structure of 4-hydroxybutyrate CoA-Transferase from Clostridium aminobutyricum | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-Hydroxybutyrate CoA-transferase, SPERMIDINE | | Authors: | Messerschmidt, A, Macieira, S, Velarde, M. | | Deposit date: | 2009-03-10 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of 4-hydroxybutyrate CoA-transferase from Clostridium aminobutyricum
Biol.Chem., 390, 2009
|
|
1SC8
 
 | | Urokinase Plasminogen Activator B-Chain-J435 Complex | | Descriptor: | N-(BENZYLSULFONYL)SERYL-N~1~-{4-[AMINO(IMINO)METHYL]BENZYL}GLYCINAMIDE, SULFATE ION, plasminogen activator, ... | | Authors: | Schweinitz, A, Steinmetzer, T, Banke, I.J, Arlt, M.J.E, Stuerzebecher, A, Schuster, O, Geissler, A, Giersiefen, H, Zeslawska, E, Jacob, U, Kruger, A, Stuerzebecher, J. | | Deposit date: | 2004-02-12 | | Release date: | 2004-06-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of novel and selective inhibitors of urokinase-type plasminogen activator with improved pharmacokinetic properties for use as antimetastatic agents
J.Biol.Chem., 279, 2004
|
|
3TC9
 
 | |
4DDL
 
 | | PDE10a Crystal Structure Complexed with Novel Inhibitor | | Descriptor: | 2-{1-[5-(6,7-dimethoxycinnolin-4-yl)-3-methylpyridin-2-yl]piperidin-4-yl}propan-2-ol, SULFATE ION, ZINC ION, ... | | Authors: | Chmait, S, Jordan, S, Zhang, J. | | Deposit date: | 2012-01-18 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of potent, selective, and metabolically stable 4-(pyridin-3-yl)cinnolines as novel phosphodiesterase 10A (PDE10A) inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
4ME8
 
 | |
6W46
 
 | | Valine-Containing Collagen Peptide | | Descriptor: | 1,2-ETHANEDIOL, Collagen-like peptide | | Authors: | Chenoweth, D.M, Melton, S.D. | | Deposit date: | 2020-03-10 | | Release date: | 2020-08-12 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Rules for the design of aza-glycine stabilized triple-helical collagen peptides.
Chem Sci, 11, 2020
|
|
4MEO
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 2-methyl-quinoline ligand | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, DIMETHYL SULFOXIDE, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Martin, S, Fedorov, O, Vidler, L.R, Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Hoelder, S, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-08-27 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Discovery of Novel Small-Molecule Inhibitors of BRD4 Using Structure-Based Virtual Screening.
J.Med.Chem., 56, 2013
|
|
2C5Y
 
 | | DIFFERENTIAL BINDING OF INHIBITORS TO ACTIVE AND INACTIVE CDK2 PROVIDES INSIGHTS FOR DRUG DESIGN | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, HYDROXY(OXO)(3-{[(2Z)-4-[3-(1H-1,2,4-TRIAZOL-1-YLMETHYL)PHENYL]PYRIMIDIN-2(5H)-YLIDENE]AMINO}PHENYL)AMMONIUM | | Authors: | Kontopidis, G, McInnes, C, Pandalaneni, S.R, McNae, I, Gibson, D, Mezna, M, Thomas, M, Wood, G, Wang, S, Walkinshaw, M.D, Fischer, P.M. | | Deposit date: | 2005-11-03 | | Release date: | 2006-03-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Differential Binding of Inhibitors to Active and Inactive Cdk2 Provides Insights for Drug Design.
Chem.Biol., 13, 2006
|
|
6CL6
 
 | | Structure of P. aeruginosa R2 pyocin fiber PA0620 comprising C-terminal residues 323-691 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, FE (III) ION, ... | | Authors: | Buth, S.A, Shneider, M.M, Leiman, P.G. | | Deposit date: | 2018-03-01 | | Release date: | 2018-06-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Structure and Analysis of R1 and R2 Pyocin Receptor-Binding Fibers.
Viruses, 10, 2018
|
|
2Y3A
 
 | | Crystal structure of p110beta in complex with icSH2 of p85beta and the drug GDC-0941 | | Descriptor: | 2-(1H-indazol-4-yl)-6-{[4-(methylsulfonyl)piperazin-1-yl]methyl}-4-morpholin-4-yl-thieno[3,2-d]pyrimidine, PHOSPHATIDYLINOSITOL 3-KINASE REGULATORY SUBUNIT BETA, PHOSPHATIDYLINOSITOL-4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT BETA ISOFORM | | Authors: | Zhang, X, Vadas, O, Perisic, O, Williams, R.L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-03-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of Lipid Kinase P110Beta-P85Beta Elucidates an Unusual Sh2-Domain-Mediated Inhibitory Mechanism.
Mol.Cell, 41, 2011
|
|
5VP8
 
 | | I38T mutant of 2009 H1N1 PA Endonuclease | | Descriptor: | 1-[(3R,5S,7R)-1,5,7,9-tetrakis(2-oxopyrrolidin-1-yl)nonan-3-yl]-1,3-dihydro-2H-pyrrol-2-one, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-05-04 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of the I38T PA Substitution as a Resistance Marker for Next-Generation Influenza Virus Endonuclease Inhibitors.
MBio, 9, 2018
|
|
