8SKR
 
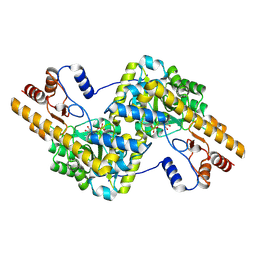 | | human liver mitochondrial Aspartate aminotransferase | | Descriptor: | Aspartate aminotransferase, mitochondrial, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Zhang, Z, Tringides, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology.
Mol.Cell Proteomics, 22, 2023
|
|
8SKQ
 
 | |
8SKP
 
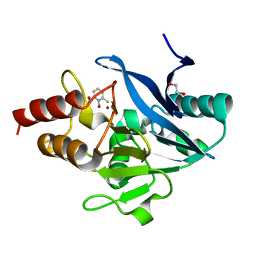 | | X-ray structure of the NDM-4 beta-lactamase from Klebsiella pneumonia in complex with 1-hydroxypyridine-2(1H)-thione-6-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 1-hydroxy-6-sulfanylidene-1,6-dihydropyridine-2-carboxylic acid, Metallo-beta-lactamase type 2, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of a novel inhibitor for the New Delhi metallo-beta-lactamase-4: Implications for drug design and combating bacterial drug resistance.
J.Biol.Chem., 299, 2023
|
|
8SKO
 
 | |
8SKN
 
 | | Crystal structure of compound 3-bound human Dynamin-1-like protein GTPase-BSE fusion | | Descriptor: | 1,2-ETHANEDIOL, Dynamin-1-like protein GTPase-BSE fusion, N-[4-(azetidin-1-yl)-2-(4-methylphenyl)quinolin-6-yl]-2-methylpropanamide | | Authors: | Ma, B. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Discovery of Potent Allosteric DRP1 Inhibitors by Disrupting Protein-Protein Interaction with MiD49.
Acs Med.Chem.Lett., 14, 2023
|
|
8SKM
 
 | |
8SKL
 
 | | PTP1B in complex with 182 | | Descriptor: | 1,2-ETHANEDIOL, 5-[1-fluoro-3-hydroxy-7-(3-hydroxy-3-methylbutoxy)naphthalen-2-yl]-1lambda~6~,2,5-thiadiazolidine-1,1,3-trione, CHLORIDE ION, ... | | Authors: | Babon, J.J, Chen, H, Tiganis, T. | | Deposit date: | 2023-04-20 | | Release date: | 2023-08-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A small molecule inhibitor of PTP1B and PTPN2 enhances T cell anti-tumor immunity.
Nat Commun, 14, 2023
|
|
8SKK
 
 | | Crystal Structure of the Tick Evasin EVA-AAM1001(L39P) Complexed to Human Chemokine CCL17 | | Descriptor: | C-C motif chemokine 17, Evasin P1243 | | Authors: | Devkota, S.R, Bhusal, R.P, Aryal, P, Wilce, M.C.J, Stone, M.J. | | Deposit date: | 2023-04-19 | | Release date: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Tick Evasin EVA-AAM1001(L39P) Complexed to Human Chemokine CCL7
To be Published
|
|
8SKJ
 
 | | Crystal structure of a Nanobody bound to the V5 peptide. | | Descriptor: | NbA1, V5 Epitope Tag Peptide | | Authors: | Zaghal, M, Matte, K, Venes, A, Patel, S, Laroche, G, Sarvan, S, Joshi, M, Couture, J.F, Giguere, P.M. | | Deposit date: | 2023-04-19 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Development of a V5-tag-directed nanobody and its implementation as an intracellular biosensor of GPCR signaling.
J.Biol.Chem., 299, 2023
|
|
8SKI
 
 | |
8SKH
 
 | | Co-structure of SARS-CoV-2 (COVID-19 with covalent pyrazoline based inhibitors | | Descriptor: | 2-chloro-1-[(4R,5R)-3,4,5-triphenyl-4,5-dihydro-1H-pyrazol-1-yl]ethan-1-one, 3C-like proteinase nsp5 | | Authors: | Knapp, M.S, Ornelas, E. | | Deposit date: | 2023-04-19 | | Release date: | 2023-06-07 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Discovery of Potent Pyrazoline-Based Covalent SARS-CoV-2 Main Protease Inhibitors.
Chembiochem, 24, 2023
|
|
8SKF
 
 | |
8SKE
 
 | |
8SKD
 
 | |
8SKB
 
 | | Crystal Structure of GDP-mannose 3,5 epimerase de Myrciaria dubia in complex with NAD | | Descriptor: | GDP-mannose 3,5 epimerase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Santillan, J.A.V, Cabrejos, D.A.L, Pereira, H.M, Gomez, J.C.C, Garratt, R.C. | | Deposit date: | 2023-04-19 | | Release date: | 2024-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural insights into the Smirnoff-Wheeler pathway for vitamin C production in the Amazon fruit camu-camu.
J.Exp.Bot., 75, 2024
|
|
8SK8
 
 | |
8SK7
 
 | | Cryo-EM structure of designed Influenza HA binder, HA_20, bound to Influenza HA (Strain: Iowa43) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HA_20 minibinder (RFdiffusion-designed), ... | | Authors: | Borst, A.J, Bennett, N.R. | | Deposit date: | 2023-04-18 | | Release date: | 2023-06-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | De novo design of protein structure and function with RFdiffusion.
Nature, 620, 2023
|
|
8SK6
 
 | | human liver mitochondrial Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase | | Descriptor: | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial | | Authors: | Zhang, Z, Tringides, M. | | Deposit date: | 2023-04-18 | | Release date: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | High-Resolution Structural Proteomics of Mitochondria Using the 'Build and Retrieve' Methodology.
Mol.Cell Proteomics, 22, 2023
|
|
8SK5
 
 | | Crystal structure of the SARS-CoV-2 neutralizing VHH 7A9 bound to the spike receptor binding domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, anti-SARS-CoV-2 receptor binding domain VHH | | Authors: | Noland, C.L, Pande, K, Zhang, L, Zhou, H, Galli, J, Eddins, M, Gomez-Llorente, Y. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Discovery and multimerization of cross-reactive single-domain antibodies against SARS-like viruses to enhance potency and address emerging SARS-CoV-2 variants.
Sci Rep, 13, 2023
|
|
8SK4
 
 | |
8SK2
 
 | |
8SK0
 
 | | Crystal structure of EvdS6 decarboxylase in ligand bound state | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Sharma, P, Frigo, L, Dulin, C.C, Bachmann, B.O, Iverson, T.M. | | Deposit date: | 2023-04-18 | | Release date: | 2023-08-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | EvdS6 is a bifunctional decarboxylase from the everninomicin gene cluster.
J.Biol.Chem., 299, 2023
|
|
8SJY
 
 | |
8SJX
 
 | |
8SJW
 
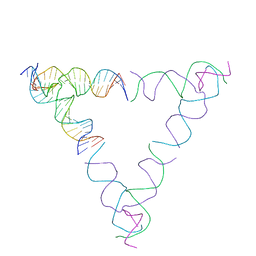 | | [4T28] Self-assembling right-handed tensegrity triangle with 28 interjunction base pairs and R3 symmetry | | Descriptor: | DNA (28-MER), DNA (5'-D(*GP*AP*AP*CP*TP*GP*CP*CP*TP*GP*AP*AP*TP*TP*AP*CP*TP*GP*AP*CP*CP*G)-3'), DNA (5'-D(*TP*CP*AP*TP*CP*AP*GP*TP*GP*GP*CP*AP*GP*T)-3'), ... | | Authors: | Janowski, J, Vecchioni, S, Sha, R, Ohayon, Y.P. | | Deposit date: | 2023-04-18 | | Release date: | 2024-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (7.64 Å) | | Cite: | Engineering tertiary chirality in helical biopolymers.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
