2HBL
 
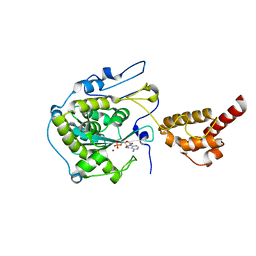 | | Structure of the yeast nuclear exosome component, Rrp6p, reveals an interplay between the active site and the HRDC domain; Protein in complex with Mn, Zn, and AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Exosome complex exonuclease RRP6, MANGANESE (II) ION, ... | | Authors: | Midtgaard, S.F, Assenholt, J, Jonstrup, A.T, Van, L.B, Jensen, T.H, Brodersen, D.E. | | Deposit date: | 2006-06-14 | | Release date: | 2006-07-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
5BUU
 
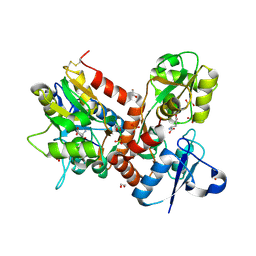 | | Crystal structure of the GluA2 ligand-binding domain (L483Y-N754S) in complex with glutamate and BPAM-321 at 2.07 A resolution | | Descriptor: | (3R)-7-chloro-2,3,4-trimethyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide, 1,2-ETHANEDIOL, GLUTAMIC ACID, ... | | Authors: | Larsen, A.P, Tapken, D, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-06-04 | | Release date: | 2016-02-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Synthesis and Pharmacology of Mono-, Di-, and Trialkyl-Substituted 7-Chloro-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-Dioxides Combined with X-ray Structure Analysis to Understand the Unexpected Structure-Activity Relationship at AMPA Receptors.
Acs Chem Neurosci, 7, 2016
|
|
2H1V
 
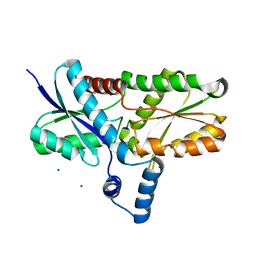 | | Crystal structure of the Lys87Ala mutant variant of Bacillus subtilis ferrochelatase | | Descriptor: | Ferrochelatase, MAGNESIUM ION | | Authors: | Hansson, M.D, Karlberg, T, Arys Rahardja, M, Al-Karadaghi, S, Hansson, M. | | Deposit date: | 2006-05-17 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Amino Acid Residues His183 and Glu264 in Bacillus subtilis Ferrochelatase Direct and Facilitate the Insertion of Metal Ion into Protoporphyrin IX
Biochemistry, 46, 2007
|
|
3D83
 
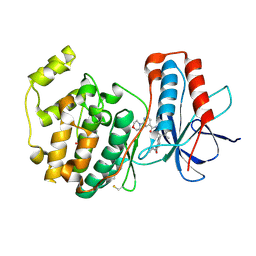 | | Crystal structure of P38 kinase in complex with a biphenyl amide inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 14, N-{4'-[(cyclopropylmethyl)carbamoyl]-6-methylbiphenyl-3-yl}-2-morpholin-4-ylpyridine-4-carboxamide | | Authors: | Somers, D.O. | | Deposit date: | 2008-05-22 | | Release date: | 2008-07-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biphenyl amide p38 kinase inhibitors 4: DFG-in and DFG-out binding modes.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3CHO
 
 | | Crystal structure of leukotriene a4 hydrolase in complex with 2-amino-N-[4-(phenylmethoxy)phenyl]-acetamide | | Descriptor: | ACETATE ION, Leukotriene A-4 hydrolase, N-[4-(benzyloxy)phenyl]glycinamide, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
3CHP
 
 | | Crystal structure of leukotriene a4 hydrolase in complex with (3S)-3-amino-4-oxo-4-[(4-phenylmethoxyphenyl)amino]butanoic acid | | Descriptor: | (3S)-3-amino-4-oxo-4-[(4-phenylmethoxyphenyl)amino]butanoic acid, ACETATE ION, IMIDAZOLE, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
3D4J
 
 | | Crystal structure of Human mevalonate diphosphate decarboxylase | | Descriptor: | Diphosphomevalonate decarboxylase, SULFATE ION | | Authors: | Voynova, N.E, Fu, Z, Battaile, K, Herdendorf, T.J, Kim, J.-J.P, Miziorko, H.M. | | Deposit date: | 2008-05-14 | | Release date: | 2008-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human mevalonate diphosphate decarboxylase: characterization, investigation of the mevalonate diphosphate binding site, and crystal structure.
Arch.Biochem.Biophys., 480, 2008
|
|
2HM7
 
 | |
3NNW
 
 | | Crystal structure of P38 alpha in complex with DP802 | | Descriptor: | 2-[3-(3-tert-butyl-5-{[(2,3-dichlorophenyl)carbamoyl]imino}-2,5-dihydro-1H-pyrazol-1-yl)phenyl]acetamide, Mitogen-activated protein kinase 14 | | Authors: | Abendroth, J. | | Deposit date: | 2010-06-24 | | Release date: | 2010-09-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Switch control pocket inhibitors of p38-MAP kinase. Durable type II inhibitors that do not require binding into the canonical ATP hinge region
Bioorg.Med.Chem.Lett., 20, 2010
|
|
4PO7
 
 | | Structure of the Sortilin:neurotensin complex at excess neurotensin concentration | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neurotensin/neuromedin N, Sortilin, ... | | Authors: | Quistgaard, E.M, Groftehauge, M.K, Thirup, S.S. | | Deposit date: | 2014-02-25 | | Release date: | 2014-07-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Revisiting the structure of the Vps10 domain of human sortilin and its interaction with neurotensin.
Protein Sci., 23, 2014
|
|
3DHZ
 
 | |
3DTG
 
 | |
2OZQ
 
 | | Crystal Structure of apo-MUP | | Descriptor: | CADMIUM ION, Novel member of the major urinary protein (Mup) gene family, SODIUM ION | | Authors: | Dennis, C.A, Homans, S.W, Phillips, S.E.V, Syme, N.R. | | Deposit date: | 2007-02-27 | | Release date: | 2008-01-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Origin of heat capacity changes in a "nonclassical" hydrophobic interaction.
Chembiochem, 8, 2007
|
|
4QDL
 
 | | Crystal structure of E.coli Cas1-Cas2 complex | | Descriptor: | CRISPR-associated endonuclease Cas1, CRISPR-associated endoribonuclease Cas2 | | Authors: | Tamulaitiene, G, Sinkunas, T, Silanskas, A, Gasiunas, G, Grazulis, S, Siksnys, V. | | Deposit date: | 2014-05-14 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of E.coli Cas1-Cas2 complex
To be Published
|
|
4QF9
 
 | | Structure of GluK1 ligand-binding domain (S1S2) in complex with (S)-2-amino-4-(2,3-dioxo-1,2,3,4-tetrahydroquinoxalin-6-yl)butanoic acid at 2.28 A resolution | | Descriptor: | (2S)-2-amino-4-(2,3-dioxo-1,2,3,4-tetrahydroquinoxalin-6-yl)butanoic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Kristensen, C.M, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2014-05-20 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Binding Mode of an alpha-Amino Acid-Linked Quinoxaline-2,3-dione Analogue at Glutamate Receptor Subtype GluK1.
ACS Chem Neurosci, 6, 2015
|
|
3ECN
 
 | | Crystal structure of PDE8A catalytic domain in complex with IBMX | | Descriptor: | 3-ISOBUTYL-1-METHYLXANTHINE, High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ... | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
3EFX
 
 | | Novel binding site identified in a hybrid between cholera toxin and heat-labile enterotoxin, 1.9A crystal structure reveals the details | | Descriptor: | Cholera enterotoxin subunit B, Heat-labile enterotoxin B chain, alpha-L-fucopyranose-(1-2)-[2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)]beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]beta-D-glucopyranose | | Authors: | Holmner, A, Lebens, M, Teneberg, S, Angstrom, J, Okvist, M, Krengel, U. | | Deposit date: | 2008-09-10 | | Release date: | 2008-09-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel binding site identified in a hybrid between cholera toxin and heat-labile enterotoxin: 1.9 A crystal structure reveals the details
Structure, 12, 2004
|
|
4P3A
 
 | |
4PC3
 
 | | Elongation factor Tu:Ts complex with partially bound GDP | | Descriptor: | Elongation factor Ts, Elongation factor Tu 1, GLYCEROL, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8313 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
2Q3J
 
 | | Crystal structure of the His183Ala variant of Bacillus subtilis ferrochelatase in complex with N-Methyl Mesoporphyrin | | Descriptor: | Ferrochelatase, MAGNESIUM ION, N-METHYL PROTOPORPHYRIN IX 2,4-DISULFONIC ACID | | Authors: | Karlberg, T, Thorvaldsen, O.H, Al-Karadaghi, S. | | Deposit date: | 2007-05-30 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Porphyrin binding and distortion and substrate specificity in the ferrochelatase reaction: the role of active site residues
J.Mol.Biol., 378, 2008
|
|
4P2Y
 
 | |
1YO5
 
 | | Analysis of the 2.0A crystal structure of the protein-DNA complex of human PDEF Ets domain bound to the prostate specific antigen regulatory site | | Descriptor: | Enhancer site of Prostate Specific Antigen Promoter Region, SAM pointed domain containing ets transcription factor | | Authors: | Wang, Y, Feng, L, Said, M, Balderman, S, Fayazi, Z, Liu, Y, Ghosh, D, Gulick, A.M. | | Deposit date: | 2005-01-26 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the 2.0 A Crystal Structure of the Protein-DNA Complex of the Human PDEF Ets Domain Bound to the Prostate Specific Antigen Regulatory Site
Biochemistry, 44, 2005
|
|
3ECM
 
 | | Crystal structure of the unliganded PDE8A catalytic domain | | Descriptor: | High affinity cAMP-specific and IBMX-insensitive 3',5'-cyclic phosphodiesterase 8A, MAGNESIUM ION, ZINC ION | | Authors: | Wang, H, Yan, Z, Yang, S, Cai, J, Robinson, H, Ke, H. | | Deposit date: | 2008-09-01 | | Release date: | 2008-11-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic and structural studies of phosphodiesterase-8A and implication on the inhibitor selectivity
Biochemistry, 47, 2008
|
|
4PC7
 
 | | Elongation factor Tu:Ts complex in a near GTP conformation. | | Descriptor: | (1S,2S,3E,5E,7E,10S,11S,12S)-12-[(2R,4E,6E,8Z,10R,12E,14E,16Z,18S,19Z)-10,18-DIHYDROXY-12,16,19-TRIMETHYL-11,22-DIOXOOX ACYCLODOCOSA-4,6,8,12,14,16,19-HEPTAEN-2-YL]-2,11-DIHYDROXY-1,10-DIMETHYL-9-OXOTRIDECA-3,5,7-TRIEN-1-YL 6-DEOXY-2,4-DI-O-METHYL-BETA-L-GALACTOPYRANOSIDE, Elongation factor Ts, Elongation factor Tu 1, ... | | Authors: | Thirup, S.S. | | Deposit date: | 2014-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.6003 Å) | | Cite: | Structural outline of the detailed mechanism for elongation factor Ts-mediated guanine nucleotide exchange on elongation factor Tu.
J.Struct.Biol., 191, 2015
|
|
1ZZL
 
 | | Crystal structure of P38 with triazolopyridine | | Descriptor: | 6-[4-(4-FLUOROPHENYL)-1,3-OXAZOL-5-YL]-3-ISOPROPYL[1,2,4]TRIAZOLO[4,3-A]PYRIDINE, Mitogen-activated protein kinase 14 | | Authors: | McClure, K.F, Han, S. | | Deposit date: | 2005-06-14 | | Release date: | 2005-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Theoretical and Experimental Design of Atypical Kinase Inhibitors: Application to p38 MAP Kinase.
J.Med.Chem., 48, 2005
|
|
