5T4I
 
 | |
5SV1
 
 | | Structure of the ExbB/ExbD complex from E. coli at pH 4.5 | | Descriptor: | Biopolymer transport protein ExbB, Biopolymer transport protein ExbD, MERCURY (II) ION | | Authors: | Celia, H, Botos, I, Lloubes, R, Buchanan, S.K, Noinaj, N. | | Deposit date: | 2016-08-04 | | Release date: | 2016-09-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural insight into the role of the Ton complex in energy transduction.
Nature, 538, 2016
|
|
5T86
 
 | | Crystal structure of CDI complex from E. coli A0 34/86 | | Descriptor: | ACETATE ION, CdiA toxin, CdiI immunity protein | | Authors: | Michalska, K, Stols, L, Jedrzejczak, R, Hayes, C.S, Goulding, C.W, Joachimiak, A, Structure-Function Analysis of Polymorphic CDI Toxin-Immunity Protein Complexes (UC4CDI), Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2016-09-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of CDI complex from E. coli A0 34/86
To Be Published
|
|
5TA1
 
 | | Crystal structure of BuGH86wt | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5T5C
 
 | |
5TCL
 
 | | Thaumatin from 4.96 A wavelength data collection | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Aurelius, O, Duman, R, El Omari, K, Mykhaylyk, V, Wagner, A. | | Deposit date: | 2016-09-15 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.201 Å) | | Cite: | Long-wavelength macromolecular crystallography - First successful native SAD experiment close to the sulfur edge.
Nucl Instrum Methods Phys Res B, 411, 2017
|
|
5SYO
 
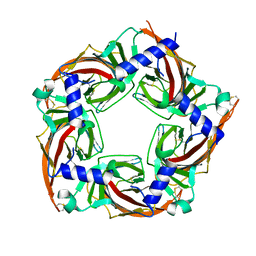 | | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with Cytisine | | Descriptor: | (1R,5S)-1,2,3,4,5,6-HEXAHYDRO-8H-1,5-METHANOPYRIDO[1,2-A][1,5]DIAZOCIN-8-ONE, Soluble acetylcholine receptor, Neuronal acetylcholine receptor subunit alpha-3 chimera | | Authors: | Bobango, J, Wu, J, Talley, I.T, Talley, T.T. | | Deposit date: | 2016-08-11 | | Release date: | 2016-10-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a chimeric acetylcholine binding protein from Aplysia californica (Ac-AChBP) containing loop C from the human alpha 3 nicotinic acetylcholine receptor in complex with Cytisine
To Be Published
|
|
5T2V
 
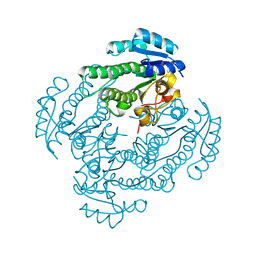 | | Crystal structure of MSMEG_6753 a putative betaketoacyl-ACP reductase | | Descriptor: | MAGNESIUM ION, Oxidoreductase, short chain dehydrogenase/reductase family protein | | Authors: | Blaise, M, Van Wyk, N, Baneres-Roquet, F, Guerardel, Y, Kremer, L. | | Deposit date: | 2016-08-24 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding of NADP(+) triggers an open-to-closed transition in a mycobacterial FabG beta-ketoacyl-ACP reductase.
Biochem. J., 474, 2017
|
|
7L04
 
 | |
5T2U
 
 | | short chain dehydrogenase/reductase family protein | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, short chain dehydrogenase/reductase family protein | | Authors: | Mickael, B, Van Wyk, N, Baneres-Roquet, F, Yann, G, Laurent, K. | | Deposit date: | 2016-08-24 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of NADP(+) triggers an open-to-closed transition in a mycobacterial FabG beta-ketoacyl-ACP reductase.
Biochem. J., 474, 2017
|
|
5T8R
 
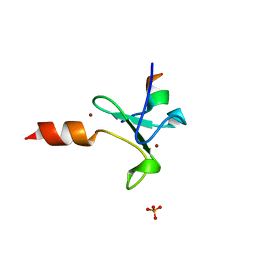 | | Crystal structure of human BAZ2A PHD zinc finger in complex with unmodified H3 10-mer | | Descriptor: | Bromodomain adjacent to zinc finger domain protein 2A, GLYCEROL, Histone H3.1, ... | | Authors: | Amato, A, Gadd, M.S, Bortoluzzi, A, Ciulli, A. | | Deposit date: | 2016-09-08 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of molecular recognition of helical histone H3 tail by PHD finger domains.
Biochem. J., 474, 2017
|
|
5SZS
 
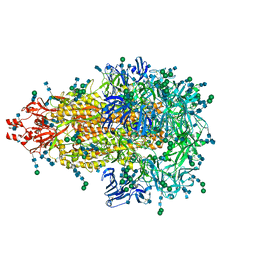 | | Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Walls, A.C, Tortorici, M.A, Frenz, B, Snijder, J, Li, W, Rey, F.A, DiMaio, F, Bosch, B.J, Veesler, D. | | Deposit date: | 2016-08-15 | | Release date: | 2016-09-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7KJZ
 
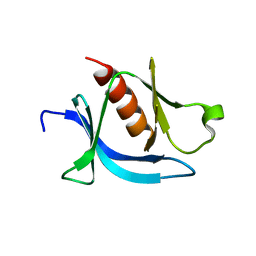 | | crystal structure of PLEKHA7 PH domain biding inositol-tetraphosphate | | Descriptor: | 1,2-ETHANEDIOL, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, Pleckstrin homology domain-containing family A member 7 | | Authors: | Marassi, F.M, Aleshin, A.E, Liddington, R.C. | | Deposit date: | 2020-10-26 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural basis for the association of PLEKHA7 with membrane-embedded phosphatidylinositol lipids.
Structure, 29, 2021
|
|
5T5K
 
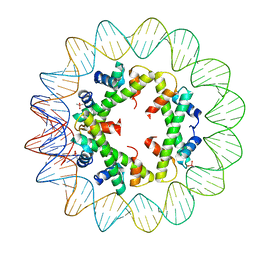 | | Structure of histone-based chromatin in Archaea | | Descriptor: | CACODYLATE ION, DNA (90-MER), DNA-binding protein HMf-2 | | Authors: | Bhattacharyya, S, Mattiroli, F, Dyer, P.N, Sandman, K, Reeve, J.N, Luger, K. | | Deposit date: | 2016-08-31 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Structure of histone-based chromatin in Archaea.
Science, 357, 2017
|
|
5T2P
 
 | | Hepatitis B virus core protein Y132A mutant in complex with sulfamoylbenzamide (SBA_R01) | | Descriptor: | 4-fluoranyl-3-(4-oxidanylpiperidin-1-yl)sulfonyl-~{N}-[3,4,5-tris(fluoranyl)phenyl]benzamide, CHLORIDE ION, Core protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2016-08-24 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.693 Å) | | Cite: | Heteroaryldihydropyrimidine (HAP) and Sulfamoylbenzamide (SBA) Inhibit Hepatitis B Virus Replication by Different Molecular Mechanisms.
Sci Rep, 7, 2017
|
|
5TF8
 
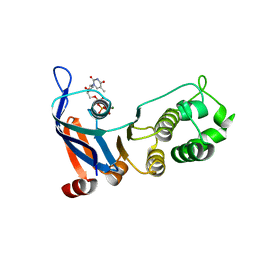 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with dTTP | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-24 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.861 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TFJ
 
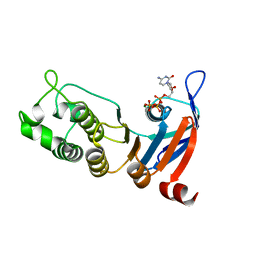 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TFF
 
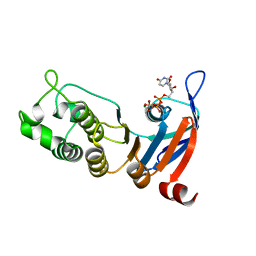 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with UTP | | Descriptor: | Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION, URIDINE 5'-TRIPHOSPHATE | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-25 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Thermodynamic correction of F508del-CFTR by ligand binding to a remote site in the mutated domain
To Be Published
|
|
5TGK
 
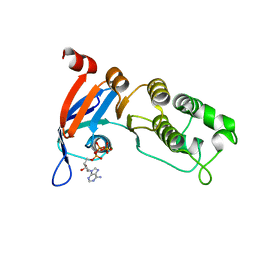 | | Nucleotide-binding domain 1 of the human cystic fibrosis transmembrane conductance regulator (CFTR) with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Wang, C, Aleksandrov, A.A, Yang, Z, Forouhar, F, Proctor, E, Kota, P, An, J, Kaplan, A, Khazanov, N, Boel, G, Stockwell, B.R, Senderowitz, H, Dokholyan, N.V, Riordan, J.R, Brouillette, C.G, Hunt, J.F. | | Deposit date: | 2016-09-28 | | Release date: | 2018-05-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | Ligand binding to a remote site thermodynamically corrects the F508del mutation in the human cystic fibrosis transmembrane conductance regulator.
J. Biol. Chem., 2018
|
|
5T63
 
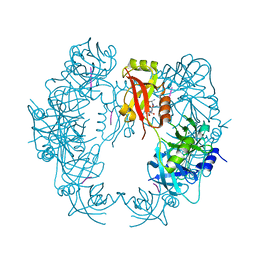 | | The HhoA protease from Synechocystis sp. PCC 6803 | | Descriptor: | ALA-ALA-ALA-ALA, MAGNESIUM ION, Putative serine protease HhoA | | Authors: | Persson, K, Hall, M, Funk, C. | | Deposit date: | 2016-09-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The HhoA protease from Synechocystis sp. PCC 6803 - Novel insights into structure and activity regulation.
J. Struct. Biol., 198, 2017
|
|
5T69
 
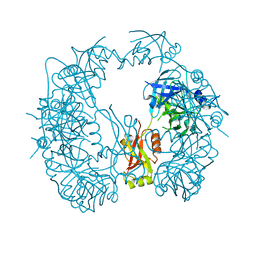 | | The HhoA protease from Synechocystis sp. PCC 6803, active site mutant | | Descriptor: | ACETATE ION, MAGNESIUM ION, Putative serine protease HhoA | | Authors: | Persson, K, Hall, M, Funk, C. | | Deposit date: | 2016-09-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The HhoA protease from Synechocystis sp. PCC 6803 - Novel insights into structure and activity regulation.
J. Struct. Biol., 198, 2017
|
|
5T94
 
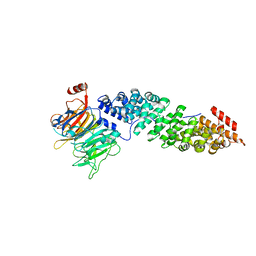 | | Crystal structure of Kap60 bound to yeast RCC1 (Prp20) | | Descriptor: | Guanine nucleotide exchange factor SRM1, Importin subunit alpha | | Authors: | Sankhala, R.S, Lokareddy, R.K, Pumroy, R.A, Cingolani, G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | Three-dimensional context rather than NLS amino acid sequence determines importin alpha subtype specificity for RCC1.
Nat Commun, 8, 2017
|
|
5TAD
 
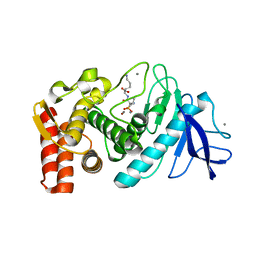 | |
5TBP
 
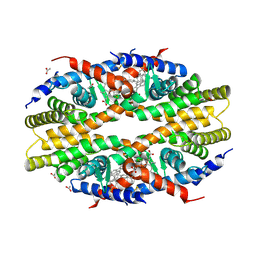 | | Crystal Structure of RXR-alpha ligand binding domain complexed with synthetic modulator K8003 | | Descriptor: | ACETATE ION, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Aleshin, A.E, Liddington, R.C, Su, Y, Zhang, X. | | Deposit date: | 2016-09-12 | | Release date: | 2017-08-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Modulation of nongenomic activation of PI3K signalling by tetramerization of N-terminally-cleaved RXR alpha.
Nat Commun, 8, 2017
|
|
5TFW
 
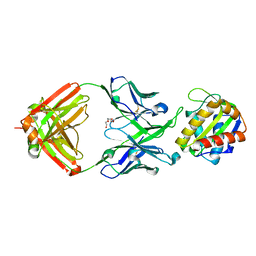 | | Crystal structure of 10E8 Fab light chain mutant2 against the MPER region of the HIV-1 Env, in complex with T117v2 epitope scaffold | | Descriptor: | 1,2-ETHANEDIOL, 10E8 EPITOPE SCAFFOLD T117V2, Antibody 10E8 FAB HEAVY CHAIN, ... | | Authors: | Irimia, A, Wilson, I.A. | | Deposit date: | 2016-09-26 | | Release date: | 2017-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.168 Å) | | Cite: | Lipid interactions and angle of approach to the HIV-1 viral membrane of broadly neutralizing antibody 10E8: Insights for vaccine and therapeutic design.
PLoS Pathog., 13, 2017
|
|
