4YLQ
 
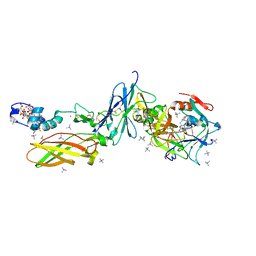 | | Crystal Structure of a FVIIa-Trypsin Chimera (FT) in Complex with Soluble Tissue Factor | | Descriptor: | CALCIUM ION, Coagulation factor VII, N-PROPANOL, ... | | Authors: | Sorensen, A.B, Svensson, L.A, Gandhi, P.S. | | Deposit date: | 2015-03-05 | | Release date: | 2015-12-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Molecular Basis of Enhanced Activity in Factor VIIa-Trypsin Variants Conveys Insights into Tissue Factor-mediated Allosteric Regulation of Factor VIIa Activity.
J.Biol.Chem., 291, 2016
|
|
4YMB
 
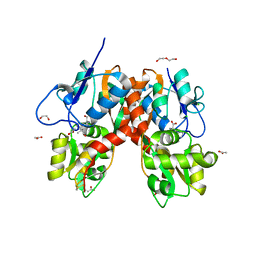 | | Structure of the ligand-binding domain of GluK1 in complex with the antagonist CNG10111 | | Descriptor: | (3R,4S)-3-(3-carboxyphenyl)-4-propyl-L-proline, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Moller, C, Tapken, D, Kastrup, J.S, Frydenvang, K. | | Deposit date: | 2015-03-06 | | Release date: | 2015-08-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure-Activity Relationship Study of Ionotropic Glutamate Receptor Antagonist (2S,3R)-3-(3-Carboxyphenyl)pyrrolidine-2-carboxylic Acid.
J.Med.Chem., 58, 2015
|
|
4Y6M
 
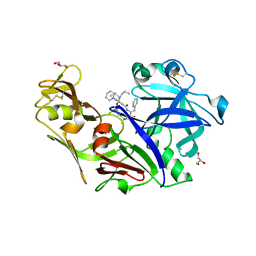 | | Structure of plasmepsin II from Plasmodium falciparum complexed with inhibitor PG418 | | Descriptor: | GLYCEROL, Plasmepsin-2, ~{N}1-[(~{Z},3~{R})-4-[2-(3-methoxyphenyl)propan-2-ylamino]-3-oxidanyl-1-phenyl-but-1-en-2-yl]-5-piperidin-1-yl-~{N}3,~{N}3-dipropyl-benzene-1,3-dicarboxamide | | Authors: | Recacha, R, Akopjana, I, Tars, K, Jaudzems, K. | | Deposit date: | 2015-02-13 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structures of plasmepsin II from Plasmodium falciparum in complex with two hydroxyethylamine-based inhibitors.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
6DHN
 
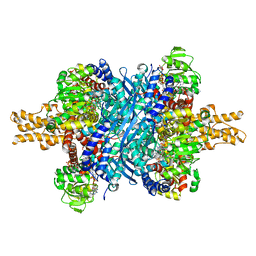 | | Bovine glutamate dehydrogenase complexed with Eu3+ | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Smith, T.J. | | Deposit date: | 2018-05-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A novel mechanism of V-type zinc inhibition of glutamate dehydrogenase results from disruption of subunit interactions necessary for efficient catalysis.
FEBS J., 278, 2011
|
|
1W7H
 
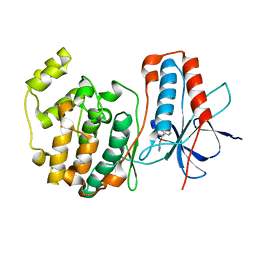 | | p38 Kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | 3-(BENZYLOXY)PYRIDIN-2-AMINE, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Jhoti, H, Gill, A, Cleasby, A, Devine, L. | | Deposit date: | 2004-09-02 | | Release date: | 2005-02-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.214 Å) | | Cite: | Fragment-Based Lead Discovery Using X-Ray Crystallography
J.Med.Chem., 48, 2005
|
|
4YA8
 
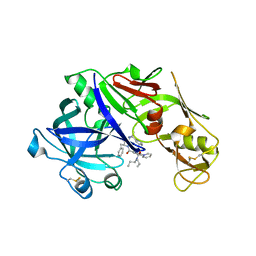 | | structure of plasmepsin II from Plasmodium Falciparum complexed with inhibitor PG394 | | Descriptor: | GLYCEROL, N'-[(2S,3S)-3-hydroxy-1-phenyl-4-{[2-(pyridin-2-yl)propan-2-yl]amino}butan-2-yl]-N,N-dipropyl-5-(pyridin-1(2H)-yl)benzene-1,3-dicarboxamide, Plasmepsin-2 | | Authors: | Recacha, R, Leitans, J, Tars, K, Jaudzems, K. | | Deposit date: | 2015-02-17 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.301 Å) | | Cite: | Structures of plasmepsin II from Plasmodium falciparum in complex with two hydroxyethylamine-based inhibitors.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4DD0
 
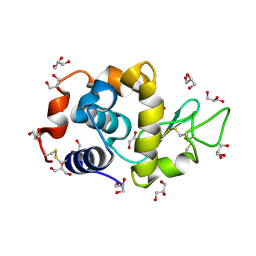 | | EVAL processed HEWL, cisplatin aqueous glycerol | | Descriptor: | GLYCEROL, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4X2C
 
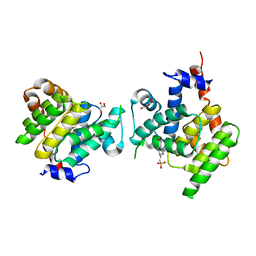 | | Clostridium difficile Fic protein_0569 mutant S31A, E35A | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Fic family protein putative filamentation induced by cAMP protein, GLYCEROL, ... | | Authors: | Jorgensen, R, Dedic, E. | | Deposit date: | 2014-11-26 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Clostridium difficile Fic_0569 S31A, E35A mutant at 1.8 Angstroms resolution
To Be Published
|
|
4X52
 
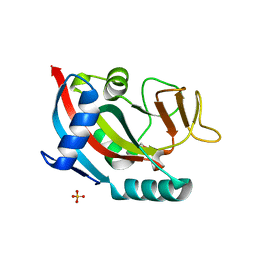 | | Human PARP13 (ZC3HAV1), C-Terminal PARP Domain (H810N; N830Y variant) | | Descriptor: | GLYCEROL, SULFATE ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Karlberg, T, Thorsell, A.G, Klepsch, M, Schuler, H. | | Deposit date: | 2014-12-04 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural Basis for Lack of ADP-ribosyltransferase Activity in Poly(ADP-ribose) Polymerase-13/Zinc Finger Antiviral Protein.
J.Biol.Chem., 290, 2015
|
|
4WFR
 
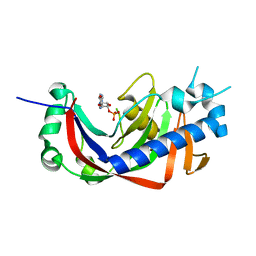 | | Catalytic domain of mouse 2',3'-cyclic nucleotide 3'- phosphodiesterase, with mutation T232A, complexed with 2'-AMP | | Descriptor: | 2',3'-cyclic-nucleotide 3'-phosphodiesterase, ADENOSINE-2'-MONOPHOSPHATE | | Authors: | Myllykoski, M, Raasakka, A, Kursula, P. | | Deposit date: | 2014-09-17 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determinants of ligand binding and catalytic activity in the myelin enzyme 2',3'-cyclic nucleotide 3'-phosphodiesterase.
Sci Rep, 5, 2015
|
|
4YWT
 
 | |
4DD3
 
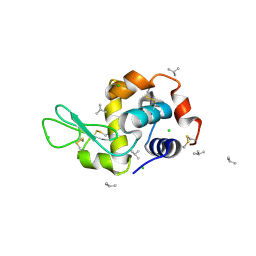 | | EVAL processed HEWL, carboplatin aqueous paratone | | Descriptor: | 2-methylprop-1-ene, CHLORIDE ION, Lysozyme C | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4DD9
 
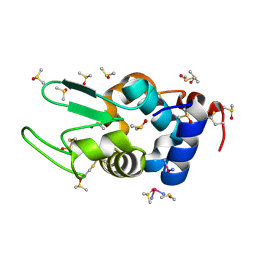 | | EVAL processed HEWL, carboplatin DMSO paratone | | Descriptor: | DIMETHYL SULFOXIDE, Lysozyme C, carboplatin | | Authors: | Tanley, S.W, Schreurs, A.M, Kroon-Batenburg, L.M, Meredith, J, Prendergast, R, Walsh, D, Bryant, P, Levy, C, Helliwell, J.R. | | Deposit date: | 2012-01-18 | | Release date: | 2012-04-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural studies of the effect that dimethyl sulfoxide (DMSO) has on cisplatin and carboplatin binding to histidine in a protein.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1WBT
 
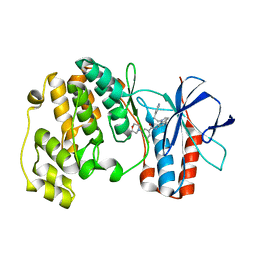 | | Identification of novel p38 alpha MAP Kinase inhibitors using fragment-based lead generation. | | Descriptor: | 3-FLUORO-5-MORPHOLIN-4-YL-N-[1-(2-PYRIDIN-4-YLETHYL)-1H-INDOL-6-YL]BENZAMIDE, MITOGEN-ACTIVATED PROTEIN KINASE 14 | | Authors: | Tickle, J, Cleasby, A, Devine, L.A, Jhoti, H. | | Deposit date: | 2004-11-05 | | Release date: | 2005-11-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of Novel P38Alpha Map Kinase Inhibitors Using Fragment-Based Lead Generation.
J.Med.Chem., 48, 2005
|
|
5DPN
 
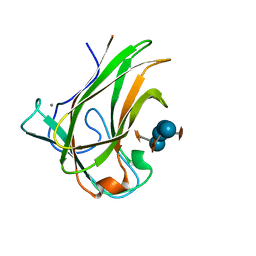 | | Engineered CBM X-2 L110F in complex with branched carbohydrate XXXG. | | Descriptor: | CALCIUM ION, Xylanase, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Ohlin, M. | | Deposit date: | 2015-09-13 | | Release date: | 2015-10-28 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.6 Å), X-RAY DIFFRACTION | | Cite: | Neutron Crystallographic Studies Reveal Hydrogen Bond and Water-Mediated Interactions between a Carbohydrate-Binding Module and Its Bound Carbohydrate Ligand.
Biochemistry, 54, 2015
|
|
5E94
 
 | |
5ECD
 
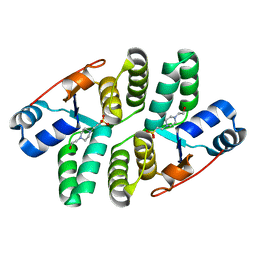 | | Structure of the Shigella flexneri VapC mutant D98N crystal form 1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, tRNA(fMet)-specific endonuclease VapC | | Authors: | Xu, K. | | Deposit date: | 2015-10-20 | | Release date: | 2016-02-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural analysis of the active site architecture of the VapC toxin from Shigella flexneri.
Proteins, 84, 2016
|
|
5DWC
 
 | |
9HE8
 
 | |
5FHO
 
 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with (S)-2-Amino-3-(5-(2-(3-chlorobenzyl)-2H-tetrazol-5-yl)-3-hydroxyisoxazol-4-yl)propanoic acid at 2.3 A resolution | | Descriptor: | (1S)-1-carboxy-2-(5-{2-[(3-chlorophenyl)methyl]-2H-tetrazol-5-yl}-3-oxo-2,3-dihydro-1,2-oxazol-4-yl)ethan-1-aminium, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tweaking Subtype Selectivity and Agonist Efficacy at (S)-2-Amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl)propionic acid (AMPA) Receptors in a Small Series of BnTetAMPA Analogues.
J.Med.Chem., 59, 2016
|
|
3MPT
 
 | | Crystal structure of P38 kinase in complex with a pyrrole-2-carboxamide inhibitor | | Descriptor: | GLYCEROL, Mitogen-activated protein kinase 14, N-(furan-2-ylmethyl)-4-[(2-methylphenyl)carbonyl]-1H-pyrrole-2-carboxamide | | Authors: | Somers, D.O. | | Deposit date: | 2010-04-27 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The discovery and initial optimisation of pyrrole-2-carboxamides as inhibitors of p38alpha MAP kinase.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
9FNM
 
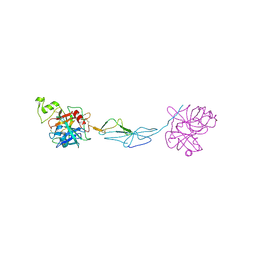 | | Structure of human haptoglobin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Haptoglobin beta chain, Isoform 2 of Haptoglobin alpha chain | | Authors: | Torvund-Jensen, M, Andersen, C.B.F. | | Deposit date: | 2024-06-10 | | Release date: | 2025-02-12 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | The Cryo-EM structure of human CD163 bound to haptoglobin-hemoglobin reveals molecular mechanisms of hemoglobin scavenging.
Nat Commun, 15, 2024
|
|
5FOQ
 
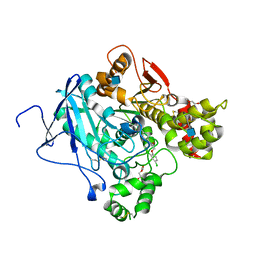 | | Acetylcholinesterase in complex with C7653 | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2,4-dichlorophenoxy)-N-[4-(1-piperidinylmethyl)phenyl]acetamide, 2-(2-METHOXYETHOXY)ETHANOL, ... | | Authors: | Berg, L, Mishra, B.K, Andersson, D.C, Ekstrom, F, Linusson, A. | | Deposit date: | 2015-11-25 | | Release date: | 2016-03-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Nature of Activated Non-Classical Hydrogen Bonds: A Case Study on Acetylcholinesterase-Ligand Complexes.
Chemistry, 22, 2016
|
|
5FHM
 
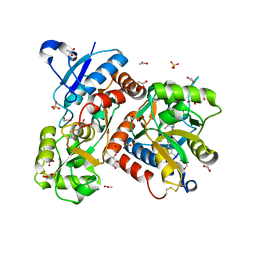 | | Crystal structure of the GluA2 ligand-binding domain (S1S2J) in complex with (S)-2-Amino-3-(5-(2-(3-(aminomethyl)benzyl)-2H-tetrazol-5-yl)-3-hydroxyisoxazol-4-yl)propanoic acid at resolution 1.55 A resolution | | Descriptor: | (2~{S})-3-[5-[2-[[3-(aminomethyl)phenyl]methyl]-1,2,3,4-tetrazol-5-yl]-3-oxidanyl-1,2-oxazol-4-yl]-2-azanyl-propanoic acid, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Kastrup, J.S, Frydenvang, K, Al-musaed, A. | | Deposit date: | 2015-12-22 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Tweaking Subtype Selectivity and Agonist Efficacy at (S)-2-Amino-3-(3-hydroxy-5-methyl-isoxazol-4-yl)propionic acid (AMPA) Receptors in a Small Series of BnTetAMPA Analogues.
J.Med.Chem., 59, 2016
|
|
5ED0
 
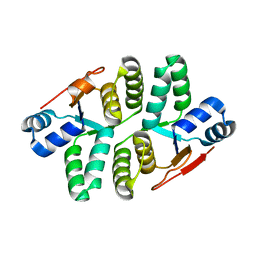 | |
