2N29
 
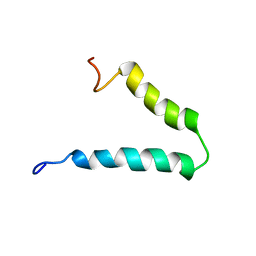 | | Solution-state NMR structure of Vpu cytoplasmic domain | | Descriptor: | Protein Vpu | | Authors: | Zhang, H, Lin, E.C, Tian, Y, Das, B.B, Opella, S.J. | | Deposit date: | 2015-05-01 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural determination of virus protein U from HIV-1 by NMR in membrane environments.
Biochim.Biophys.Acta, 1848, 2015
|
|
3NEE
 
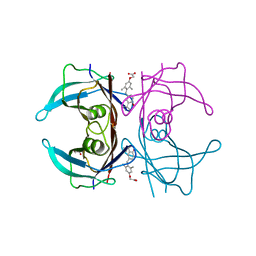 | | Wild type human transthyretin (TTR) complexed with GC-1 (TTRwt:GC-1) | | Descriptor: | GLYCEROL, Transthyretin, {4-[4-hydroxy-3-(1-methylethyl)benzyl]-3,5-dimethylphenoxy}acetic acid | | Authors: | Trivella, D.B.B, Polikarpov, I. | | Deposit date: | 2010-06-08 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The binding of synthetic triiodo l-thyronine analogs to human transthyretin: molecular basis of cooperative and non-cooperative ligand recognition.
J.Struct.Biol., 173, 2011
|
|
1HBO
 
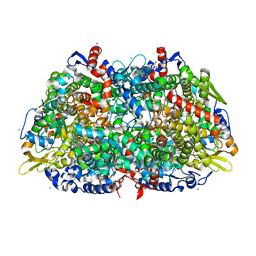 | | METHYL-COENZYME M REDUCTASE MCR-RED1-SILENT | | Descriptor: | 1-THIOETHANESULFONIC ACID, CHLORIDE ION, Coenzyme B, ... | | Authors: | Grabarse, W. | | Deposit date: | 2001-04-20 | | Release date: | 2001-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | On the Mechanism of Biological Methane Formation: Structural Evidence for Conformational Changes in Methyl-Coenzyme M Reductase Upon Substrate Binding
J.Mol.Biol., 309, 2001
|
|
2XFY
 
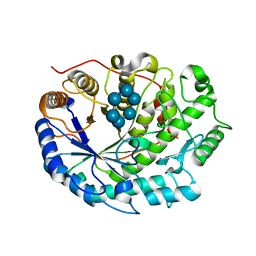 | | Crystal structure of Barley Beta-Amylase complexed with alpha- cyclodextrin | | Descriptor: | 1,2-ETHANEDIOL, BETA-AMYLASE, Cyclohexakis-(1-4)-(alpha-D-glucopyranose) | | Authors: | Rejzek, M, Stevenson, C.E.M, Southard, A.M, Stanley, D, Denyer, K, Smith, A.M, Naldrett, M.J, Lawson, D.M, Field, R.A. | | Deposit date: | 2010-05-28 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.207 Å) | | Cite: | Chemical Genetics and Cereal Starch Metabolism: Structural Basis of the Non-Covalent and Covalent Inhibition of Barley Beta-Amylase.
Mol.Biosyst., 7, 2011
|
|
5QF9
 
 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with compound_FMOMB000242a | | Descriptor: | 1-(2,6-dihydroxy-3-propylphenyl)ethan-1-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Keedy, D.A, Hill, Z.B, Biel, J.T, Kang, E, Rettenmaier, T.J, Brandao-Neto, J, von Delft, F, Wells, J.A, Fraser, J.S. | | Deposit date: | 2018-08-30 | | Release date: | 2018-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | An expanded allosteric network in PTP1B by multitemperature crystallography, fragment screening, and covalent tethering.
Elife, 7, 2018
|
|
3NES
 
 | | V30M mutant human transthyretin (TTR) complexed with GC-1 (V30M:GC-1) | | Descriptor: | GLYCEROL, Transthyretin, {4-[4-hydroxy-3-(1-methylethyl)benzyl]-3,5-dimethylphenoxy}acetic acid | | Authors: | Trivella, D.B.B, Polikarpov, I. | | Deposit date: | 2010-06-09 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The binding of synthetic triiodo l-thyronine analogs to human transthyretin: molecular basis of cooperative and non-cooperative ligand recognition.
J.Struct.Biol., 173, 2011
|
|
5QQJ
 
 | | PanDDA analysis group deposition -- Crystal Structure of Kalirin/Rac1 in complex with MolPort-009-587-558 | | Descriptor: | 1,2-ETHANEDIOL, Kalirin, N-{1-[(imidazo[1,2-a]pyridin-2-yl)methyl]-1H-pyrazol-4-yl}-N'-(5-methyl-1,2-oxazol-3-yl)urea, ... | | Authors: | Gray, J.L, Krojer, T, Talon, R, Douangamath, A, Jimenez Antunez, C, Bountra, C, Arrowsmith, C.H, Edwards, A, Brennan, P.E, von Delft, F. | | Deposit date: | 2019-05-03 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
4HA4
 
 | | Structure of beta-glycosidase from Acidilobus saccharovorans in complex with glycerol | | Descriptor: | 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, Beta-galactosidase, GLYCEROL, ... | | Authors: | Trofimov, A.A, Polyakov, K.M, Tikhonov, A.V, Bezsudnova, E.Y, Dorovatovskii, P.V, Gumerov, V.M, Ravin, N.V, Skryabin, K.G, Popov, V.O. | | Deposit date: | 2012-09-25 | | Release date: | 2012-10-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structures of beta-glycosidase from Acidilobus saccharovorans in complexes with tris and glycerol.
Dokl.Biochem.Biophys., 449
|
|
1HFN
 
 | | NMR solution structures of vMIP-II 1-71 from Kaposi's sarcoma-associated herpesvirus. | | Descriptor: | VIRAL MACROPHAGE INFLAMMATORY PROTEIN-II | | Authors: | Crump, M.P, Elisseeva, E, Gong, J.-H, Clark-Lewis, I, Sykes, B.D. | | Deposit date: | 2000-12-07 | | Release date: | 2001-01-07 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Structure/Function of Human Herpesvirus-8 Mip-II (1-71) and the Antagonist N-Terminal Segment (1-10)
FEBS Lett., 489, 2001
|
|
2C31
 
 | | CRYSTAL STRUCTURE OF OXALYL-COA DECARBOXYLASE IN COMPLEX WITH THE COFACTOR DERIVATIVE THIAMIN-2-THIAZOLONE DIPHOSPHATE AND ADENOSINE DIPHOSPHATE | | Descriptor: | 2-{3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-4-METHYL-2-OXO-2,3-DIHYDRO-1,3-THIAZOL-5-YL}ETHYL TRIHYDROGEN DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Berthold, C.L, Moussatche, P, Richards, N.G.J, Lindqvist, Y. | | Deposit date: | 2005-10-03 | | Release date: | 2005-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural Basis for Activation of the Thiamin Diphosphate-Dependent Enzyme Oxalyl-Coa Decarboxylase by Adenosine Diphosphate.
J.Biol.Chem., 280, 2005
|
|
5QOY
 
 | | PanDDA analysis group deposition -- Crystal Structure of DCP2 (NUDT20) in complex with YW-FY-378 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(2H-1,3-benzodioxole-5-carbonyl)-2,3,4,5-tetrahydro-1H-1,4-diazepin-1-yl]ethan-1-one, ACETATE ION, ... | | Authors: | Nelson, E.R, Velupillai, S, Talon, R, Collins, P.M, Krojer, T, Wang, D, Brandao-Neto, J, Douangamath, A, Burgess-Brown, N, Arrowsmith, C.H, Bountra, C, Huber, K, von Delft, F. | | Deposit date: | 2019-02-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1FSG
 
 | | TOXOPLASMA GONDII HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE COMPLEXED WITH 9-DEAZAGUANINE, ALPHA-D-5-PHOSPHORIBOSYL-1-PYROPHOSPHATE (PRPP) AND TWO MG2+ IONS | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, 9-DEAZAGUANINE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE, ... | | Authors: | Heroux, A, White, E.L, Ross, L.J, Kuzin, A.P, Borhani, D.W. | | Deposit date: | 2000-09-08 | | Release date: | 2000-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Substrate deformation in a hypoxanthine-guanine phosphoribosyltransferase ternary complex: the structural basis for catalysis.
Structure Fold.Des., 8, 2000
|
|
4D8C
 
 | | Crystal Structure of Human Beta Secretase in Complex with NVP-BXD552, derived from a co-crystallization experiment | | Descriptor: | (3S,4S,5R)-3-(4-amino-3-{[(2R)-3-ethoxy-1,1,1-trifluoropropan-2-yl]oxy}-5-fluorobenzyl)-5-[(3-tert-butylbenzyl)amino]tetrahydro-2H-thiopyran-4-ol 1,1-dioxide, Beta-secretase 1, SULFATE ION | | Authors: | Rondeau, J.M, Bourgier, E. | | Deposit date: | 2012-01-10 | | Release date: | 2012-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of cyclic sulfone hydroxyethylamines as potent and selective beta-site APP-cleaving enzyme 1 (BACE1) inhibitors: structure based design and in vivo reduction of amyloid beta-peptides
J.Med.Chem., 55, 2012
|
|
3CWB
 
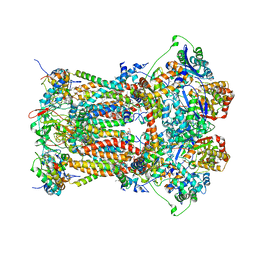 | | Chicken Cytochrome BC1 Complex inhibited by an iodinated analogue of the polyketide Crocacin-D | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, AZIDE ION, CARDIOLIPIN, ... | | Authors: | Huang, L, Cromartie, T, Viner, R, Crowley, P.J, Berry, E.A. | | Deposit date: | 2008-04-21 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | The role of molecular modeling in the design of analogues of the fungicidal natural products crocacins A and D.
Bioorg.Med.Chem., 16, 2008
|
|
4G27
 
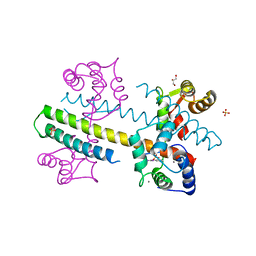 | | Calcium-calmodulin complexed with the calmodulin binding domain from a small conductance potassium channel splice variant and phenylurea | | Descriptor: | 1-phenylurea, CALCIUM ION, Calmodulin, ... | | Authors: | Zhang, M, Pascal, J.M, Zhang, J.-F. | | Deposit date: | 2012-07-11 | | Release date: | 2012-09-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Identification of the functional binding pocket for compounds targeting small-conductance Ca(2+)-activated potassium channels.
Nat Commun, 3, 2012
|
|
2MYS
 
 | |
3ST8
 
 | |
2XUK
 
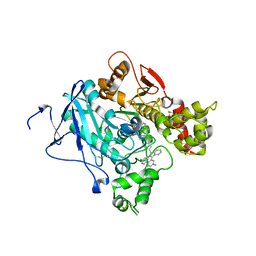 | | CRYSTAL STRUCTURE OF MACHE-Y337A-TZ2PA6 SYN COMPLEX (10 MTH) | | Descriptor: | 3,8-DIAMINO-6-PHENYL-5-[6-[1-[2-[(1,2,3,4-TETRAHYDRO-9-ACRIDINYL)AMINO]ETHYL]-1H-1,2,3-TRIAZOL-5-YL]HEXYL]-PHENANTHRIDINIUM, ACETYLCHOLINESTERASE | | Authors: | Bourne, Y, Radic, Z, Taylor, P, Marchot, P. | | Deposit date: | 2010-10-19 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformational Remodeling of Femtomolar Inhibitor-Acetylcholinesterase Complexes in the Crystalline State
J.Am.Chem.Soc., 132, 2010
|
|
4EY2
 
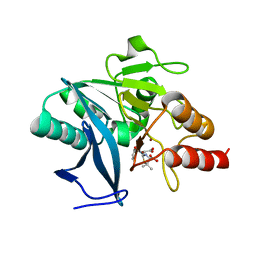 | | Crystal structure of NDM-1 bound to hydrolyzed methicillin | | Descriptor: | (2R,4S)-2-{(R)-carboxy[(2,6-dimethoxybenzoyl)amino]methyl}-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, Beta-lactamase NDM-1, ZINC ION | | Authors: | Strynadka, N.C.J, King, D.T. | | Deposit date: | 2012-05-01 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | New Delhi Metallo-Beta-Lactamase: Structural Insights into Beta-Lactam Recognition and Inhibition
J.Am.Chem.Soc., 134, 2012
|
|
4EYL
 
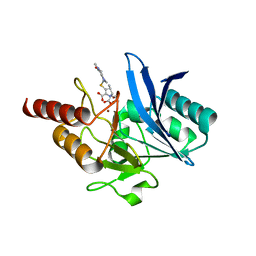 | | Crystal structure of NDM-1 bound to hydrolyzed meropenem | | Descriptor: | (2S)-2-[(1S,2R)-1-carboxy-2-hydroxypropyl]-4-{[(3S,5S)-5-(dimethylcarbamoyl)pyrrolidin-3-yl]sulfanyl}-3-methyl-2H-pyrro le-5-carboxylic acid, Beta-lactamase NDM-1, ZINC ION | | Authors: | Strynadka, N.C.J, King, D.T. | | Deposit date: | 2012-05-01 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Delhi Metallo-Beta-Lactamase: Structural Insights into Beta-Lactam Recognition and Inhibition
J.Am.Chem.Soc., 134, 2012
|
|
2DCX
 
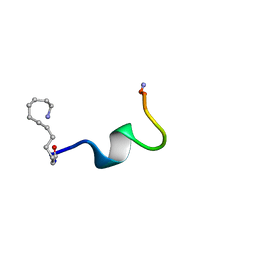 | | NMR solution structure of the Dermaseptin antimicrobial peptide analog NC12-K4S4(1-13)a | | Descriptor: | 12-AMINO-DODECANOIC ACID, Dermaseptin-4 | | Authors: | Shalev, D.E, Rotem, S, Fish, A, Mor, A. | | Deposit date: | 2006-01-17 | | Release date: | 2006-02-28 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Consequences of N-acylation on structure and membrane binding properties of dermaseptin derivative k4-s4-(1-13)
J.Biol.Chem., 281, 2006
|
|
2DD6
 
 | | Solution structure of Dermaseptin antimicrobial peptide truncated, mutated analog, K4-S4(1-13)a | | Descriptor: | Dermaseptin-4 | | Authors: | Shalev, D.E, Rotem, S, Fish, A, Mor, A. | | Deposit date: | 2006-01-19 | | Release date: | 2006-02-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Consequences of N-acylation on structure and membrane binding properties of dermaseptin derivative k4-s4-(1-13)
J.Biol.Chem., 281, 2006
|
|
5T0A
 
 | | Crystal Structure of Heparan Sulfate 6-O-Sulfotransferase with bound PAP and heptasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, Krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
3Q25
 
 | | Crystal structure of human alpha-synuclein (1-19) fused to maltose binding protein (MBP) | | Descriptor: | GLYCEROL, Maltose-binding periplasmic protein/alpha-synuclein chimeric protein, SULFATE ION, ... | | Authors: | Zhao, M, Sawaya, M.R, Cascio, D, Eisenberg, D. | | Deposit date: | 2010-12-19 | | Release date: | 2011-06-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of segments of alpha-synuclein fused to maltose-binding protein suggest intermediate states during amyloid formation
Protein Sci., 20, 2011
|
|
3PS1
 
 | | Crystal structure of the Escherichia Coli LPXC/LPC-011 complex | | Descriptor: | 4-[4-(4-aminophenyl)buta-1,3-diyn-1-yl]-N-[(2S,3R)-3-hydroxy-1-(hydroxyamino)-1-oxobutan-2-yl]benzamide, 4-ethynyl-N-[(1S,2R)-2-hydroxy-1-(oxocarbamoyl)propyl]benzamide, DIMETHYL SULFOXIDE, ... | | Authors: | Lee, C.-J, Zhou, P. | | Deposit date: | 2010-11-30 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Syntheses, structures and antibiotic activities of LpxC inhibitors based on the diacetylene scaffold.
Bioorg.Med.Chem., 19, 2011
|
|
