7NP6
 
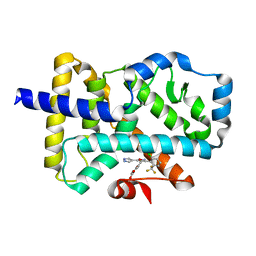 | | ROR(gamma)t ligand binding domain in complex with allosteric ligand FM257 | | Descriptor: | 4-[[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrazol-4-yl)-1,2-oxazol-4-yl]methoxy]benzoic acid, Nuclear receptor ROR-gamma | | Authors: | Oerlemans, G.J.M, Somsen, B.A, de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Activity Relationship Studies of Trisubstituted Isoxazoles as Selective Allosteric Ligands for the Retinoic-Acid-Receptor-Related Orphan Receptor gamma t.
J.Med.Chem., 64, 2021
|
|
8DFC
 
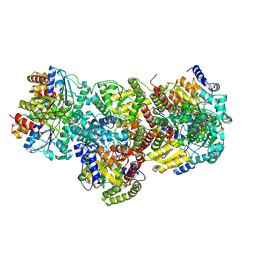 | |
1EI2
 
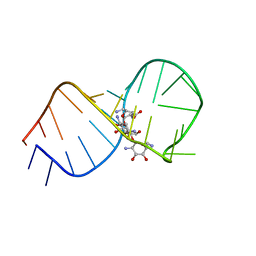 | | STRUCTURAL BASIS FOR RECOGNITION OF THE RNA MAJOR GROOVE IN THE TAU EXON 10 SPLICING REGULATORY ELEMENT BY AMINOGLYCOSIDE ANTIBIOTICS | | Descriptor: | NEOMYCIN, TAU EXON 10 SRE RNA | | Authors: | Varani, L, Spillantini, M.G, Goedert, M, Varani, G. | | Deposit date: | 2000-02-23 | | Release date: | 2000-03-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for recognition of the RNA major groove in the tau exon 10 splicing regulatory element by aminoglycoside antibiotics.
Nucleic Acids Res., 28, 2000
|
|
6M2N
 
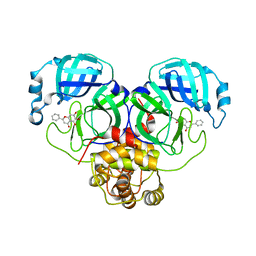 | | SARS-CoV-2 3CL protease (3CL pro) in complex with a novel inhibitor | | Descriptor: | 3C-like proteinase, 5,6,7-trihydroxy-2-phenyl-4H-chromen-4-one | | Authors: | Su, H.X, Zhao, W.F, Li, M.J, Xie, H, Xu, Y.C. | | Deposit date: | 2020-02-28 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients.
Acta Pharmacol.Sin., 41, 2020
|
|
8DFD
 
 | |
1MT9
 
 | | Viability of a drug-resistant HIV-1 protease mutant: structural insights for better antiviral therapy | | Descriptor: | PHOSPHATE ION, PROTEASE RETROPEPSIN, p1-p6 Gag substrate decapeptide | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Viability of drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy
J.Virol., 77, 2003
|
|
1MRL
 
 | | Crystal structure of streptogramin A acetyltransferase with dalfopristin | | Descriptor: | 5-(2-DIETHYLAMINO-ETHANESULFONYL)-21-HYDROXY-10-ISOPROPYL-11,19-DIMETHYL-9,26-DIOXA-3,15,28-TRIAZA-TRICYCLO[23.2.1.00,255]OCTACOSA-1(27),12,17,19,25(28)-PENTAENE-2,8,14,23-TETRAONE, Streptogramin A acetyltransferase | | Authors: | Kehoe, L.E, Snidwongse, J, Courvalin, P, Rafferty, J.B, Murray, I.A. | | Deposit date: | 2002-09-18 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Synercid (Quinupristin-Dalfopristin) Resistance in Gram-positive Bacterial Pathogens
J.Biol.Chem., 278, 2003
|
|
1E9X
 
 | |
1W7G
 
 | | Alpha-thrombin complex with sulfated hirudin (residues 54-65) and L- Arginine template inhibitor CS107 | | Descriptor: | HIRUDIN I, N-{(1S)-1-{[4-(3-AMINOPROPYL)PIPERAZIN-1-YL]CARBONYL}-4-[(DIAMINOMETHYLENE)AMINO]BUTYL}-3-(TRIFLUOROMETHYL)BENZENESULFONAMIDE, THROMBIN | | Authors: | Remiche, J, Sauvage, E, Herman, R, Charlier, P. | | Deposit date: | 2004-09-02 | | Release date: | 2006-05-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Design, Synthesis and Evaluation of Graftable Thrombin Inhibitors for the Preparation of Blood-Compatible Polymer Materials.
Org.Biomol.Chem., 3, 2005
|
|
1EF2
 
 | | CRYSTAL STRUCTURE OF MANGANESE-SUBSTITUTED KLEBSIELLA AEROGENES UREASE | | Descriptor: | MANGANESE (II) ION, UREASE ALPHA SUBUNIT, UREASE BETA SUBUNIT, ... | | Authors: | Yamaguchi, K, Cosper, N.J, Stalhandske, C, Scott, R.A, Pearson, M.A, Karplus, P.A, Hausinger, R.P. | | Deposit date: | 2000-02-05 | | Release date: | 2000-03-09 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterization of metal-substituted Klebsiella aerogenes urease.
J.Biol.Inorg.Chem., 4, 1999
|
|
1EC0
 
 | | HIV-1 protease in complex with the inhibitor bea403 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DI-2-FLUORO-BENZYL-GLUCARYL]-DI-[1-AMINO-INDAN-2-OL] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Symmetric fluoro-substituted diol-based HIV protease inhibitors. Ortho-fluorinated and meta-fluorinated P1/P1'-benzyloxy side groups significantly improve the antiviral activity and preserve binding efficacy
Eur.J.Biochem., 271, 2004
|
|
4Q9N
 
 | | Crystal structure of Chlamydia trachomatis enoyl-ACP reductase (FabI) in complex with NADH and AFN-1252 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], N-methyl-N-[(3-methyl-1-benzofuran-2-yl)methyl]-3-(7-oxo-5,6,7,8-tetrahydro-1,8-naphthyridin-3-yl)propanamide | | Authors: | Yao, J, Abdelrahman, Y, Robertson, R.M, Cox, J.V, Belland, R.J, White, S.W, Rock, C.O. | | Deposit date: | 2014-05-01 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.795 Å) | | Cite: | Type II Fatty Acid Synthesis Is Essential for the Replication of Chlamydia trachomatis.
J.Biol.Chem., 289, 2014
|
|
1W6F
 
 | | Arylamine N-acetyltransferase from Mycobacterium smegmatis with the anti-tubercular drug isoniazid bound in the active site. | | Descriptor: | 4-(DIAZENYLCARBONYL)PYRIDINE, ARYLAMINE N-ACETYLTRANSFERASE | | Authors: | Sandy, J, Holton, S, Fullham, E, Sim, E, Noble, M.E.M. | | Deposit date: | 2004-08-17 | | Release date: | 2005-02-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of the Anti-Tubercular Drug Isoniazid to the Arylamine N-Acetyltransferase Protein from Mycobacterium Smegmatis
Protein Sci., 14, 2005
|
|
4PYK
 
 | | human COMT, double domain swap | | Descriptor: | CHLORIDE ION, Catechol O-methyltransferase, MAGNESIUM ION, ... | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1EXA
 
 | | ENANTIOMER DISCRIMINATION ILLUSTRATED BY CRYSTAL STRUCTURES OF THE HUMAN RETINOIC ACID RECEPTOR HRARGAMMA LIGAND BINDING DOMAIN: THE COMPLEX WITH THE ACTIVE R-ENANTIOMER BMS270394. | | Descriptor: | DODECYL-ALPHA-D-MALTOSIDE, R-3-FLUORO-4-[2-HYDROXY-2-(5,5,8,8-TETRAMETHYL-5,6,7,8,-TETRAHYDRO-NAPHTALEN-2-YL)-ACETYLAMINO]-BENZOIC ACID, RETINOIC ACID RECEPTOR GAMMA-2 | | Authors: | Klaholz, B.P, Mitschler, A, Belema, M, Zusi, C, Moras, D, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2000-05-02 | | Release date: | 2000-06-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARgamma.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1MR9
 
 | | Crystal structure of Streptogramin A Acetyltransferase with acetyl-CoA bound | | Descriptor: | ACETYL COENZYME *A, Streptogramin A Acetyltransferase | | Authors: | Kehoe, L.E, Snidwongse, J, Courvalin, P, Rafferty, J.B, Murray, I.A. | | Deposit date: | 2002-09-18 | | Release date: | 2003-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis of Synercid (Quinupristin-Dalfopristin) Resistance in Gram-positive Bacterial Pathogens
J.Biol.Chem., 278, 2003
|
|
6M2Q
 
 | | SARS-CoV-2 3CL protease (3CL pro) apo structure (space group C21) | | Descriptor: | 3C-like proteinase | | Authors: | Su, H.X, Zhao, W.F, Li, M.J, Xie, H, Xu, Y.C. | | Deposit date: | 2020-02-28 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients.
Acta Pharmacol.Sin., 41, 2020
|
|
1EC3
 
 | | HIV-1 protease in complex with the inhibitor MSA367 | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DIBENZYL-GLUCARYL]-DI-[VALINYL-AMINOMETHANYL-PYRIDINE] | | Authors: | Unge, T. | | Deposit date: | 2000-01-25 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Optimization of P1-P3 groups in symmetric and asymmetric HIV-1 protease inhibitors
Eur.J.Biochem., 270, 2003
|
|
1EPR
 
 | | ENDOTHIA ASPARTIC PROTEINASE (ENDOTHIAPEPSIN) COMPLEXED WITH PD-135,040 | | Descriptor: | ENDOTHIAPEPSIN, N~2~-[(2R)-2-benzyl-3-(tert-butylsulfonyl)propanoyl]-N-{(1R)-1-(cyclohexylmethyl)-3,3-difluoro-2,2-dihydroxy-4-[(2-morpholin-4-ylethyl)amino]-4-oxobutyl}-3-(1H-imidazol-3-ium-4-yl)-L-alaninamide | | Authors: | Badasso, M, Crawford, M, Cooper, J.B, Blundell, T.L. | | Deposit date: | 1994-07-27 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural comparison of 21 inhibitor complexes of the aspartic proteinase from Endothia parasitica.
Protein Sci., 3, 1994
|
|
1W5X
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,3-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1S,2R)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXAN EDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
1MT8
 
 | | Viability of a drug-resistant HIV-1 protease mutant: structural insights for better antiviral therapy | | Descriptor: | ACETATE ION, Capsid-p2 substrate peptide of HIV-1 Gag polyprotein, PROTEASE RETROPEPSIN | | Authors: | Prabu-Jeyabalan, M, Nalivaika, E.A, King, N.M, Schiffer, C.A. | | Deposit date: | 2002-09-20 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Viability of drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy
J.Virol., 77, 2003
|
|
6MEL
 
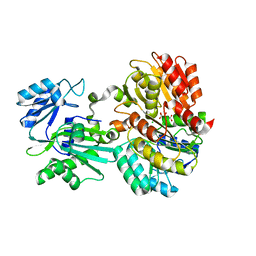 | | Succinyl-CoA synthase from Campylobacter jejuni | | Descriptor: | CHLORIDE ION, CITRIC ACID, Succinate--CoA ligase [ADP-forming] subunit beta, ... | | Authors: | Osipiuk, J, Maltseva, N, Jedrzejczak, R, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-06 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Succinyl-CoA synthase from Campylobacter jejuni
to be published
|
|
1EHJ
 
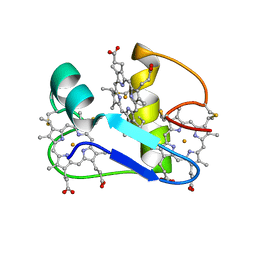 | | A PROTON-NMR INVESTIGATION OF THE FULLY REDUCED CYTOCHROME C7 FROM DESULFUROMONAS ACETOXIDANS | | Descriptor: | CYTOCHROME C7, HEME C | | Authors: | Assfalg, M, Banci, L, Bertini, I, Bruschi, M, Giudici-Orticoni, M.T. | | Deposit date: | 2000-02-21 | | Release date: | 2000-05-10 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | A proton-NMR investigation of the fully reduced cytochrome c7 from Desulfuromonas acetoxidans. Comparison between the reduced and the oxidized forms.
Eur.J.Biochem., 266, 1999
|
|
1W5V
 
 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | HIV-1 PROTEASE, N,N-[2,5-O-DI-3-FLUORO-BENZYL-GLUCARYL]-DI-[1-AMINO-INDAN-2-OL] | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
4PYM
 
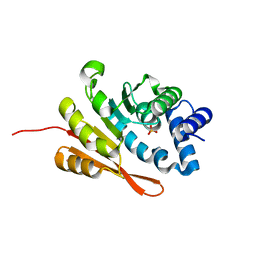 | | humanized rat apo-COMT bound to sulphate | | Descriptor: | Catechol O-methyltransferase, POTASSIUM ION, SULFATE ION | | Authors: | Ehler, A, Benz, J, Schlatter, D, Rudolph, M.G. | | Deposit date: | 2014-03-27 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Mapping the conformational space accessible to catechol-O-methyltransferase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
