5M0W
 
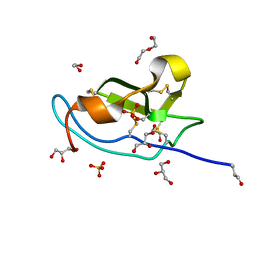 | | N-terminal domain of mouse Shisa 3 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Lohkamp, B, Ojala, J.R.M. | | Deposit date: | 2016-10-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Ab initio solution of macromolecular crystal structures without direct methods.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LWH
 
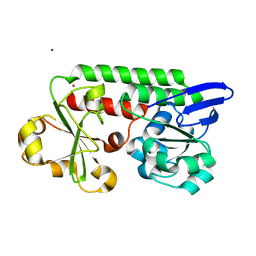 | | CeuE (Y288F variant) a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin ABC transporter substrate-binding protein, ZINC ION | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
4KMV
 
 | | Structure of the L100F MUTANT OF DEHALOPEROXIDASE-HEMOGLOBIN A FROM AMPHITRITE ORNATA WITH 2,4,6-TRICHLOROPHENOL | | Descriptor: | 1,2-ETHANEDIOL, 2,4,6-trichlorophenol, Dehaloperoxidase A, ... | | Authors: | Wang, C, Lovelace, L, Lebioda, L. | | Deposit date: | 2013-05-08 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Complexes of dual-function hemoglobin/dehaloperoxidase with substrate 2,4,6-trichlorophenol are inhibitory and indicate binding of halophenol to compound I.
Biochemistry, 52, 2013
|
|
4KN3
 
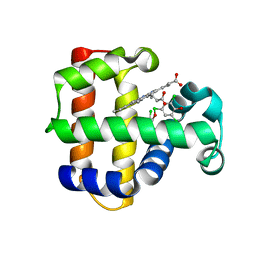 | | Structure of the Y34NS91G double mutant of Dehaloperoxidase from Amphitrite ornata with 2,4,6-trichlorophenol | | Descriptor: | 2,4,6-trichlorophenol, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, C, Lovelace, L, Lebioda, L. | | Deposit date: | 2013-05-08 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Complexes of dual-function hemoglobin/dehaloperoxidase with substrate 2,4,6-trichlorophenol are inhibitory and indicate binding of halophenol to compound I.
Biochemistry, 52, 2013
|
|
4KMW
 
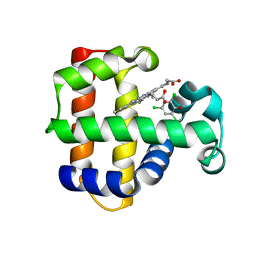 | | Structure of the Y34N MUTANT OF DEHALOPEROXIDASE-HEMOGLOBIN A FROM AMPHITRITE ORNATA WITH 2,4,6-TRICHLOROPHENOL | | Descriptor: | 2,4,6-trichlorophenol, Dehaloperoxidase A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, C, Lovelace, L, Lebioda, L. | | Deposit date: | 2013-05-08 | | Release date: | 2013-09-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Complexes of dual-function hemoglobin/dehaloperoxidase with substrate 2,4,6-trichlorophenol are inhibitory and indicate binding of halophenol to compound I.
Biochemistry, 52, 2013
|
|
3O87
 
 | | Crystal structure of AmpC beta-lactamase in complex with a sulfonamide boronic acid inhibitor | | Descriptor: | 4-({[(dihydroxyboranyl)methyl]sulfamoyl}methyl)benzoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Romagnoli, C, Karpiak, J, Shoichet, B.K. | | Deposit date: | 2010-08-02 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Design, Synthesis, Crystal Structures, and Antimicrobial Activity of Sulfonamide Boronic Acids as beta-Lactamase Inhibitors
J.Med.Chem., 53, 2010
|
|
2RIS
 
 | |
4IJH
 
 | | Fragment-based Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A | | Descriptor: | 3-chloro-6-[3-(4-fluorophenyl)-5-sulfanyl-4H-1,2,4-triazol-4-yl]-1-benzothiophene-2-carboxylic acid, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Patrone, J.D, Kennedy, J.P, Frank, A.O, Vangamudi, B, Pelz, N.F, Rossanese, O.W, Waterson, A.G, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2012-12-21 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A.
ACS MED.CHEM.LETT., 4, 2013
|
|
2RIU
 
 | | Alternative models for two crystal structures of Candida albicans 3,4-dihydroxy-2-butanone 4-phosphate synthase- alternate interpreation | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, RIBULOSE-5-PHOSPHATE | | Authors: | Stenkamp, R.E, Le Trong, I. | | Deposit date: | 2007-10-12 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Alternative models for two crystal structures of Candida albicans 3,4-dihydroxy-2-butanone 4-phosphate synthase.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3G6J
 
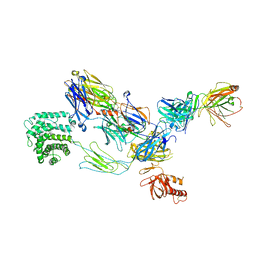 | | C3b in complex with a C3b specific Fab | | Descriptor: | CALCIUM ION, Complement C3 alpha chain, Complement C3 beta chain, ... | | Authors: | Wiesmann, C. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Functional Analysis of a C3b-specific Antibody That Selectively Inhibits the Alternative Pathway of Complement
J.Biol.Chem., 284, 2009
|
|
4JLQ
 
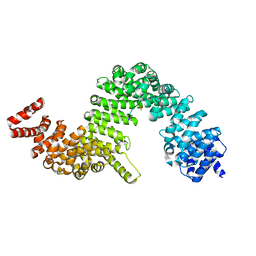 | | Crystal structure of human Karyopherin-beta2 bound to the PY-NLS of Saccharomyces cerevisiae NAB2 | | Descriptor: | Nuclear polyadenylated RNA-binding protein NAB2, Transportin-1 | | Authors: | Sampathkumar, P, Gizzi, A, Rout, M.P, Chook, Y.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of human Karyopherin beta 2 bound to the PY-NLS of Saccharomyces cerevisiae Nab2.
J.Struct.Funct.Genom., 14, 2013
|
|
5NSD
 
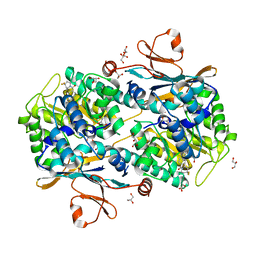 | | Co-crystal structure of NAMPT dimer with KPT-9274 | | Descriptor: | (~{E})-3-(6-azanylpyridin-3-yl)-~{N}-[[5-[4-[4,4-bis(fluoranyl)piperidin-1-yl]carbonylphenyl]-7-(4-fluorophenyl)-1-benzofuran-2-yl]methyl]prop-2-enamide, GLYCEROL, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Neggers, J.E, Kwanten, B, Dierckx, T, Noguchi, H, Voet, A, Vercruysse, T, Baloglu, E, Senapedis, W, Jacquemyn, M, Daelemans, D. | | Deposit date: | 2017-04-26 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.046 Å) | | Cite: | Target identification of small molecules using large-scale CRISPR-Cas mutagenesis scanning of essential genes.
Nat Commun, 9, 2018
|
|
4HB9
 
 | |
8TM7
 
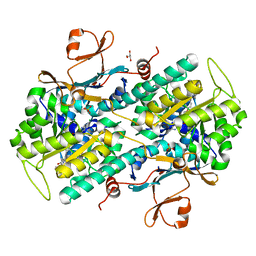 | |
4G31
 
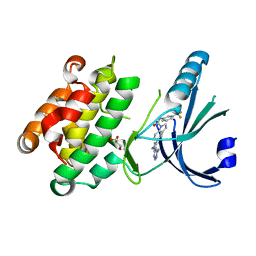 | | Crystal Structure of GSK6414 Bound to PERK (R587-R1092, delete A660-T867) at 2.28 A Resolution | | Descriptor: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-2,3-dihydro-1H-indol-1-yl]-2-[3-(trifluoromethyl)phenyl]ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3, GLYCEROL | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
3O86
 
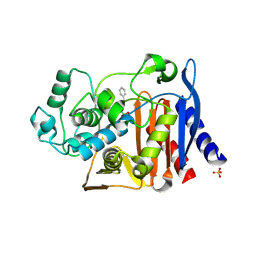 | | Crystal structure of AmpC beta-lactamase in complex with a sulfonamide boronic acid inhibitor | | Descriptor: | Beta-lactamase, PHOSPHATE ION, {[(benzylsulfonyl)amino]methyl}boronic acid | | Authors: | Eidam, O, Romagnoli, C, Karpiak, J, Shoichet, B.K. | | Deposit date: | 2010-08-02 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design, Synthesis, Crystal Structures, and Antimicrobial Activity of Sulfonamide Boronic Acids as beta-Lactamase Inhibitors
J.Med.Chem., 53, 2010
|
|
1BCX
 
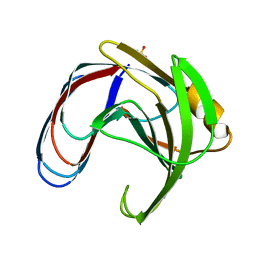 | |
4IJL
 
 | | Fragment-based Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit, {[5-(3-chloro-1-benzothiophen-2-yl)-4-phenyl-4H-1,2,4-triazol-3-yl]sulfanyl}acetic acid | | Authors: | Feldkamp, M.D, Patrone, J.D, Kennedy, J.P, Frank, A.O, Vangamudi, B, Pelz, N.F, Rossanese, O.W, Waterson, A.G, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2012-12-21 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of Protein-Protein Interaction Inhibitors of Replication Protein A.
ACS MED.CHEM.LETT., 4, 2013
|
|
4G34
 
 | | Crystal Structure of GSK6924 Bound to PERK (R587-R1092, delete A660-T867) at 2.70 A Resolution | | Descriptor: | 1-[5-(4-aminothieno[3,2-c]pyridin-3-yl)-2,3-dihydro-1H-indol-1-yl]-2-phenylethanone, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of 7-Methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a Potent and Selective First-in-Class Inhibitor of Protein Kinase R (PKR)-like Endoplasmic Reticulum Kinase (PERK).
J.Med.Chem., 55, 2012
|
|
3O88
 
 | | Crystal structure of AmpC beta-lactamase in complex with a sulfonamide boronic acid inhibitor | | Descriptor: | 3-[(2R)-2-[(benzylsulfonyl)amino]-2-(dihydroxyboranyl)ethyl]benzoic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Eidam, O, Romagnoli, C, Karpiak, J, Shoichet, B.K. | | Deposit date: | 2010-08-02 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Design, Synthesis, Crystal Structures, and Antimicrobial Activity of Sulfonamide Boronic Acids as beta-Lactamase Inhibitors
J.Med.Chem., 53, 2010
|
|
4M7I
 
 | | Crystal Structure of GSK6157 Bound to PERK (R587-R1092, delete A660-T867) at 2.34A Resolution | | Descriptor: | 1-[5-(4-amino-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-5-yl)-4-fluoro-1H-indol-1-yl]-2-(6-methylpyridin-2-yl)ethanone, Eukaryotic translation initiation factor 2-alpha kinase 3 | | Authors: | Gampe, R.T, Axten, J.M. | | Deposit date: | 2013-08-12 | | Release date: | 2014-09-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Discovery of 5-{4-fluoro-1-[(6-methyl-2-pyridinyl)acetyl]-2,3-dihydro-1H-indol-5-yl}-7-methyl-7H-pyrrolo[2,3-d]pyrimidin-4-amine (GSK2656157), a Potent and Selective PERK Inhibitor Selected for Preclinical Development
To be Published
|
|
4NB3
 
 | | Crystal structure of RPA70N in complex with a 3,4 dichlorophenylalanine ATRIP derived peptide | | Descriptor: | 3,4 dichlorophenylalanine ATRIP derived peptide, Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Souza-Fagundes, E.M, Luzwik, J.W, Cortez, D, Olejniczak, O.T, Waterson, A.G, Rossanese, O.W, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-10-22 | | Release date: | 2014-02-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of a Potent Stapled Helix Peptide That Binds to the 70N Domain of Replication Protein A.
J.Med.Chem., 57, 2014
|
|
9BRX
 
 | | SARS-CoV-2 Papain-like Protease (PLpro) with Fragment 10 | | Descriptor: | (4R)-N-(2,4-dimethylphenyl)-7-methyl[1,2,4]triazolo[4,3-a]pyrimidin-5-amine, Papain-like protease nsp3, SULFATE ION, ... | | Authors: | Amporndanai, K, Zhao, B, Fesik, S.W. | | Deposit date: | 2024-05-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-Based Screen of SARS-CoV-2 Papain-like Protease (PL pro ).
Acs Med.Chem.Lett., 15, 2024
|
|
9BRW
 
 | | SARS-CoV-2 Papain-like Protease (PLpro) with Fragment 7 | | Descriptor: | CHLORIDE ION, N-(2-chlorophenyl)-1-methyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Papain-like protease nsp3, ... | | Authors: | Amporndanai, K, Zhao, B, Fesik, S.W. | | Deposit date: | 2024-05-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Screen of SARS-CoV-2 Papain-like Protease (PL pro ).
Acs Med.Chem.Lett., 15, 2024
|
|
9BRV
 
 | | SARS-CoV-2 Papain-like Protease (PLpro) with Fragment 5 | | Descriptor: | CHLORIDE ION, N-[2-(dimethylamino)ethyl]-N'-(3-methylphenyl)thiourea, Papain-like protease nsp3, ... | | Authors: | Amporndanai, K, Zhao, B, Fesik, S.W. | | Deposit date: | 2024-05-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Fragment-Based Screen of SARS-CoV-2 Papain-like Protease (PL pro ).
Acs Med.Chem.Lett., 15, 2024
|
|
