7ZIZ
 
 | | X-ray structure of the dead variant haloalkane dehalogenase HaloTag7-D106A bound to a pentanol tetramethylrhodamine ligand (TMR-Hy5) | | Descriptor: | CHLORIDE ION, GLYCEROL, Haloalkane dehalogenase, ... | | Authors: | Tarnawski, M, Kompa, J, Johnsson, K, Hiblot, J. | | Deposit date: | 2022-04-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exchangeable HaloTag Ligands for Super-Resolution Fluorescence Microscopy.
J.Am.Chem.Soc., 145, 2023
|
|
7ZJ0
 
 | | X-ray structure of the haloalkane dehalogenase HaloTag7 bound to a pentylmethanesulfonamide tetramethylrhodamine ligand (TMR-S5) | | Descriptor: | GLYCEROL, Haloalkane dehalogenase, [9-[2-carboxy-5-[2-[2-[5-(methylsulfonylamino)pentoxy]ethoxy]ethylcarbamoyl]phenyl]-6-(dimethylamino)xanthen-3-ylidene]-dimethyl-azanium | | Authors: | Tarnawski, M, Kompa, J, Johnsson, K, Hiblot, J. | | Deposit date: | 2022-04-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Exchangeable HaloTag Ligands for Super-Resolution Fluorescence Microscopy.
J.Am.Chem.Soc., 145, 2023
|
|
4DSN
 
 | | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity | | Descriptor: | 1,2-ETHANEDIOL, GTPase KRas, isoform 2B, ... | | Authors: | Oh, A, Maurer, T, Garrenton, L.S, Pitts, K, Anderson, D.J, Skelton, N.J, Fauber, B.P, Pan, B, Malek, S, Stokoe, D, Ludlam, M, Bowman, K.K, Wu, J, Giannetti, A.M, Starovasnik, M.A, Mellman, I, Jackson, P.K, Ruldolph, J, Fang, G, Wang, W. | | Deposit date: | 2012-02-19 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1UMI
 
 | | Structural basis of sugar-recognizing ubiquitin ligase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, F-box only protein 2 | | Authors: | Mizushima, T, Hirao, T, Yoshida, Y, Lee, S.J, Chiba, T, Iwai, K, Yamaguchi, Y, Kato, K, Tsukihara, T, Tanaka, K. | | Deposit date: | 2003-10-01 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of sugar-recognizing ubiquitin ligase.
Nat.Struct.Mol.Biol., 11, 2004
|
|
9EOY
 
 | | Structure of Thr354Asn, Glu355Gln, Thr412Asn, Ile414Met, Ile464His, and Phe467Met mutant human CaMKII alpha hub bound to PIPA | | Descriptor: | 2-[6-(4-chlorophenyl)imidazo[1,2-b]pyridazin-2-yl]ethanoic acid, ACETATE ION, Calcium/calmodulin-dependent protein kinase type II subunit alpha, ... | | Authors: | Narayanan, D, Larsen, A.S.G, Solbak, S.M.O, Wellendorph, P, Gee, C.L, Kastrup, J.S. | | Deposit date: | 2024-03-15 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Ligand-induced CaMKII alpha hub Trp403 flip, hub domain stacking, and modulation of kinase activity.
Protein Sci., 33, 2024
|
|
5HEW
 
 | | Pentameric ligand-gated ion channel ELIC mutant T28D | | Descriptor: | Gamma-aminobutyric-acid receptor subunit beta-1 | | Authors: | Engeler, S, Dutzler, R. | | Deposit date: | 2016-01-06 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Signal Transduction at the Domain Interface of Prokaryotic Pentameric Ligand-Gated Ion Channels.
Plos Biol., 14, 2016
|
|
2YZ1
 
 | |
5VZV
 
 | | TRIM23 RING domain | | Descriptor: | E3 ubiquitin-protein ligase TRIM23, ZINC ION | | Authors: | Pornillos, O, Dawidziak, D. | | Deposit date: | 2017-05-29 | | Release date: | 2017-08-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.812 Å) | | Cite: | Structure and catalytic activation of the TRIM23 RING E3 ubiquitin ligase.
Proteins, 85, 2017
|
|
5XCZ
 
 | | Structure of the cellobiohydrolase Cel6A from Phanerochaete chrysosporium in complex with cellobiose at 2.1 angstrom | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glucanase, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Tachioka, M, Nakamura, A, Ishida, T, Igarashi, K, Samejima, M. | | Deposit date: | 2017-03-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a family 6 cellobiohydrolase from the basidiomycete Phanerochaete chrysosporium
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3C7I
 
 | |
3LVV
 
 | | BSO-inhibited ScGCL | | Descriptor: | (2S)-2-amino-4-(S-butyl-N-phosphonosulfonimidoyl)butanoic acid, ADENOSINE-5'-DIPHOSPHATE, Glutamate--cysteine ligase, ... | | Authors: | Barycki, J.J, Biterova, E.I. | | Deposit date: | 2010-02-22 | | Release date: | 2010-03-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for feedback and pharmacological inhibition of Saccharomyces cerevisiae glutamate cysteine ligase.
J.Biol.Chem., 285, 2010
|
|
1FTJ
 
 | |
229L
 
 | | GENERATING LIGAND BINDING SITES IN T4 LYSOZYME USING DEFICIENCY-CREATING SUBSTITUTIONS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GUANIDINE, ... | | Authors: | Baldwin, E.P, Baase, W.A, Zhang, X.-J, Feher, V, Matthews, B.W. | | Deposit date: | 1997-06-26 | | Release date: | 1998-03-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Generation of ligand binding sites in T4 lysozyme by deficiency-creating substitutions.
J.Mol.Biol., 277, 1998
|
|
3BOV
 
 | | Crystal structure of the receptor binding domain of mouse PD-L2 | | Descriptor: | FORMIC ACID, Programmed cell death 1 ligand 2, SODIUM ION | | Authors: | Lazar-Molnar, E, Ramagopal, U, Cao, E, Toro, R, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-12-17 | | Release date: | 2008-07-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
1FW0
 
 | |
1P20
 
 | |
1T8A
 
 | | USE OF SEQUENCE DUPLICATION TO ENGINEER A LIGAND-TRIGGERED LONG-DISTANCE MOLECULAR SWITCH IN T4 Lysozyme | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, GUANIDINE, ... | | Authors: | Yousef, M.S, Baase, W.A, Matthews, B.W. | | Deposit date: | 2004-05-11 | | Release date: | 2004-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Use of sequence duplication to engineer a ligand-triggered, long-distance molecular switch in T4 lysozyme.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3DGO
 
 | | A non-biological ATP binding protein with a Tyr-Phe mutation in the ligand binding domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP Binding Protein-DX, CHLORIDE ION, ... | | Authors: | Simmons, C.R, Allen, J.P, Chaput, J.C. | | Deposit date: | 2008-06-13 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A synthetic protein selected for ligand binding affinity mediates ATP hydrolysis.
Acs Chem.Biol., 4, 2009
|
|
5H0I
 
 | |
5EDV
 
 | | Structure of the HOIP-RBR/UbcH5B~ubiquitin transfer complex | | Descriptor: | E3 ubiquitin-protein ligase RNF31, Polyubiquitin-B, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Lechtenberg, B.C, Mace, P.D, Sanishvili, R, Riedl, S.J. | | Deposit date: | 2015-10-22 | | Release date: | 2016-01-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Structure of a HOIP/E2~ubiquitin complex reveals RBR E3 ligase mechanism and regulation.
Nature, 529, 2016
|
|
1LDD
 
 | | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF Ubiquitin Ligase Complex | | Descriptor: | Anaphase Promoting Complex | | Authors: | Zheng, N, Schulman, B.A, Song, L, Miller, J.J, Jeffrey, P.D, Wang, P, Chu, C, Koepp, D.M, Elledge, S.J, Pagano, M, Conaway, R.C, Conaway, J.W, Harper, J.W, Pavletich, N.P. | | Deposit date: | 2002-04-08 | | Release date: | 2002-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex.
Nature, 416, 2002
|
|
4EGM
 
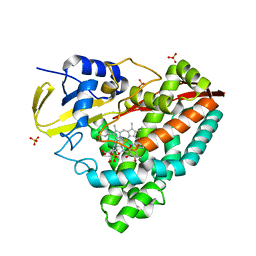 | | The X-ray crystal structure of CYP199A4 in complex with 4-ethylbenzoic acid | | Descriptor: | 4-ethylbenzoic acid, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Zhou, W, Bell, S.G, Yang, W, Zhou, R.M, Tan, A.B.H, Wong, L.-L. | | Deposit date: | 2012-03-31 | | Release date: | 2013-02-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Investigation of the substrate range of CYP199A4: modification of the partition between hydroxylation and desaturation activities by substrate and protein engineering
Chemistry, 18, 2012
|
|
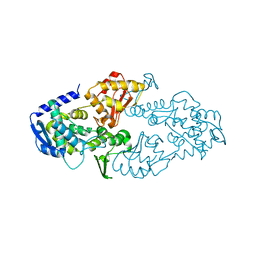
6LOH
 
 | |
6LUQ
 
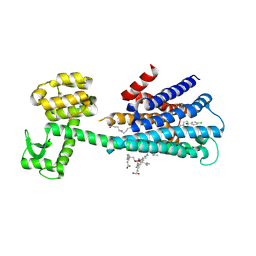 | | Haloperidol bound D2 dopamine receptor structure inspired discovery of subtype selective ligands | | Descriptor: | 4-[4-(4-chlorophenyl)-4-hydroxypiperidin-1-yl]-1-(4-fluorophenyl)butan-1-one, OLEIC ACID, chimera of D(2) dopamine receptor and Endolysin | | Authors: | Fan, L, Tan, L, Chen, Z, Qi, J, Nie, F, Luo, Z, Cheng, J, Wang, S. | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Haloperidol bound D2dopamine receptor structure inspired the discovery of subtype selective ligands.
Nat Commun, 11, 2020
|
|
3IMX
 
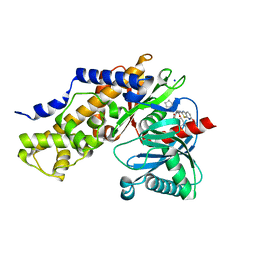 | | Crystal Structure of human glucokinase in complex with a synthetic activator | | Descriptor: | (2R)-3-cyclopentyl-N-(5-methoxy[1,3]thiazolo[5,4-b]pyridin-2-yl)-2-{4-[(4-methylpiperazin-1-yl)sulfonyl]phenyl}propanamide, Glucokinase, SODIUM ION, ... | | Authors: | Stams, T, Vash, B. | | Deposit date: | 2009-08-11 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Investigation of functionally liver selective glucokinase activators for the treatment of type 2 diabetes.
J.Med.Chem., 52, 2009
|
|
