2LGC
 
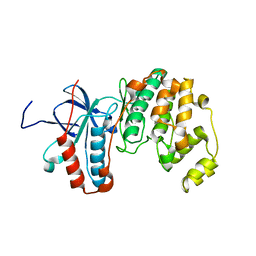 | |
4QBO
 
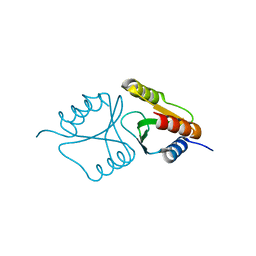 | | VRR_NUC domain | | Descriptor: | MAGNESIUM ION, Nuclease | | Authors: | Smerdon, S.J, Pennell, S, Li, J. | | Deposit date: | 2014-05-08 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | FAN1 activity on asymmetric repair intermediates is mediated by an atypical monomeric virus-type replication-repair nuclease domain.
Cell Rep, 8, 2014
|
|
4QHO
 
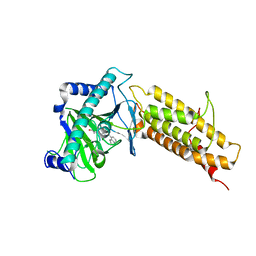 | | Crystal structure of the human fat mass and obesity associated protein (FTO) in complex with CCO10 | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase FTO, N-{[3-hydroxy-6-(naphthalen-1-yl)pyridin-2-yl]carbonyl}glycine, ZINC ION | | Authors: | Aik, W.S, Clunie-O'Connor, C, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2014-05-28 | | Release date: | 2015-06-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure-Based Design of Selective Fat Mass and Obesity Associated Protein (FTO) Inhibitors.
J.Med.Chem., 64, 2021
|
|
3TW1
 
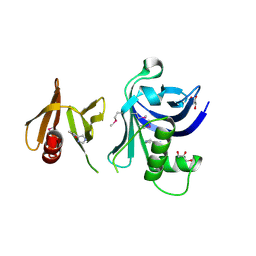 | | Structure of Rtt106-AHN | | Descriptor: | GLYCEROL, Histone chaperone RTT106, N-[2-(1H-IMIDAZOL-4-YL)ETHYL]ACETAMIDE | | Authors: | Su, D, Thompson, J.R, Mer, G. | | Deposit date: | 2011-09-21 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106.
Nature, 483, 2012
|
|
1ZN6
 
 | | X-ray Crystal Structure of Protein Q7WLM8 from Bordetella bronchiseptica. Northeast Structural Genomics Consortium Target BoR19. | | Descriptor: | SULFATE ION, phage-related conserved hypothetical protein | | Authors: | Kuzin, A.P, Yong, W, Vorobiev, S.M, Xiao, R, Ma, L.-C, Acton, T, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-05-11 | | Release date: | 2005-05-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Novel structure of the hypothetical protein Q7WLM8 from Bordetella bronchiseptica. Northeast Structural Genomics Consortium target BoR19.
To be Published
|
|
4QNQ
 
 | |
4QKN
 
 | | Crystal structure of FTO bound to a selective inhibitor | | Descriptor: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO, GLYCEROL, ... | | Authors: | Yang, C.-G, Huang, Y, Gan, J. | | Deposit date: | 2014-06-07 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5.
Nucleic Acids Res., 43, 2015
|
|
6RVV
 
 | | Structure of left-handed protein cage consisting of 24 eleven-membered ring proteins held together by gold (I) bridges. | | Descriptor: | GOLD ION, Transcription attenuation protein MtrB | | Authors: | Malay, A.D, Miyazaki, N, Biela, A.P, Iwasaki, K, Heddle, J.G. | | Deposit date: | 2019-06-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ultra-stable gold-coordinated protein cage displaying reversible assembly.
Nature, 569, 2019
|
|
4R4L
 
 | | Crystal structure of wt cGMP dependent protein kinase I alpha (PKGI alpha) leucine zipper | | Descriptor: | HEXANE-1,6-DIOL, SULFATE ION, cGMP-dependent protein kinase 1 | | Authors: | Reger, A.S, Guo, E, Yang, M.P, Qin, L, Kim, C. | | Deposit date: | 2014-08-19 | | Release date: | 2015-09-23 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.245 Å) | | Cite: | Structures of cGMP-Dependent Protein Kinase (PKG) I alpha Leucine Zippers Reveal an Interchain Disulfide Bond Important for Dimer Stability.
Biochemistry, 54, 2015
|
|
4R9T
 
 | | L-ficolin complexed to sulphates | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Laffly, E, Lacroix, M, Martin, L, Vassal-Stermann, E, Thielens, N, Gaboriaud, C. | | Deposit date: | 2014-09-08 | | Release date: | 2014-11-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Human ficolin-2 recognition versatility extended: An update on the binding of ficolin-2 to sulfated/phosphated carbohydrates.
Febs Lett., 588, 2014
|
|
6RVW
 
 | | Structure of right-handed protein cage consisting of 24 eleven-membered ring proteins held together by gold (I) bridges. | | Descriptor: | GOLD ION, Transcription attenuation protein MtrB | | Authors: | Malay, A.D, Miyazaki, N, Biela, A.P, Iwasaki, K, Heddle, J.G. | | Deposit date: | 2019-06-03 | | Release date: | 2019-06-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | An ultra-stable gold-coordinated protein cage displaying reversible assembly.
Nature, 569, 2019
|
|
4R7E
 
 | | Structure of Bre1 RING domain | | Descriptor: | E3 ubiquitin-protein ligase BRE1, ZINC ION | | Authors: | Kumar, P, Wolberger, C. | | Deposit date: | 2014-08-27 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Structure of the yeast Bre1 RING domain.
Proteins, 83, 2015
|
|
7OJ4
 
 | | Crystal structure of apo NS3 helicase from tick-borne encephalitis virus | | Descriptor: | NS3 helicase domain | | Authors: | Anindita, P.D, Havlickova, P, Kascakova, B, Grinkevich, P, Brynda, J, Franta, Z. | | Deposit date: | 2021-05-13 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.828 Å) | | Cite: | Mechanistic insight into the RNA-stimulated ATPase activity of tick-borne encephalitis virus helicase.
J.Biol.Chem., 298, 2022
|
|
4IDU
 
 | |
8A4F
 
 | | Human Interleukin-4 mutant - C3T-IL4 | | Descriptor: | Interleukin-4 | | Authors: | Vaz, D.C, Rodrigues, J.R, Mueller, T.D, Sebald, W, Redfield, C, Brito, R.M.M. | | Deposit date: | 2022-06-11 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-17 | | Method: | SOLUTION NMR | | Cite: | Lessons on protein structure from interleukin-4: All disulfides are not created equal.
Proteins, 92, 2024
|
|
6O5L
 
 | |
6OXB
 
 | |
6OYB
 
 | |
6OYE
 
 | |
4R4M
 
 | | Crystal structure of C42L cGMP dependent protein kinase I alpha (PKGI alpha) leucine zipper | | Descriptor: | SULFATE ION, cGMP-dependent protein kinase 1 | | Authors: | Reger, A.S, Guo, E, Yang, M.P, Qin, L, Kim, C. | | Deposit date: | 2014-08-19 | | Release date: | 2015-09-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.922 Å) | | Cite: | Structures of cGMP-Dependent Protein Kinase (PKG) I alpha Leucine Zippers Reveal an Interchain Disulfide Bond Important for Dimer Stability.
Biochemistry, 54, 2015
|
|
4J1X
 
 | |
6ECL
 
 | | Crystal Structure of a 1,2,4-Triazole Allosteric RNase H Inhibitor in Complex with HIV Reverse Transcriptase | | Descriptor: | 1-[4-methoxy-3-[[5-methyl-4-(phenylmethyl)-1,2,4-triazol-3-yl]sulfanylmethyl]phenyl]ethanone, MANGANESE (II) ION, Reverse transcriptase/ribonuclease H | | Authors: | Kirby, K.A, Sarafianos, S.G. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.385 Å) | | Cite: | Crystal Structure of a 1,2,4-Triazole Allosteric RNase H Inhibitor in Complex with HIV Reverse Transcriptase
To be published
|
|
4J1W
 
 | |
6OYG
 
 | |
6OY8
 
 | |
