5MGK
 
 | |
5MGT
 
 | | Complex of human NKR-P1 and LLT1 in deglycosylated forms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-type lectin domain family 2 member D, CHLORIDE ION, ... | | Authors: | Blaha, J, Skalova, T, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
6R3X
 
 | |
5MGR
 
 | | Human receptor NKR-P1 in glycosylated form, extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor subfamily B member 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Skalova, T, Blaha, J, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
6R42
 
 | |
5MGX
 
 | |
6R4A
 
 | | Aurora-A in complex with shape-diverse fragment 55 | | Descriptor: | 2-(benzimidazol-1-yl)-~{N}-(2-phenylethyl)ethanamide, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.937 Å) | | Cite: | Construction of a Shape-Diverse Fragment Set: Design, Synthesis and Screen against Aurora-A Kinase.
Chemistry, 25, 2019
|
|
5MHC
 
 | | Crystal structure of 14-3-3sigma and a p53 C-terminal 12-mer synthetic phosphopeptide | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, LYS-LEU-MET-PHE-LYS-TPO-GLU-GLY-PRO-ASP-SER-ASP, ... | | Authors: | Andrei, S, Ottmann, C, Leysen, S. | | Deposit date: | 2016-11-24 | | Release date: | 2017-10-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Small-molecule stabilization of the p53 - 14-3-3 protein-protein interaction.
FEBS Lett., 591, 2017
|
|
6R4K
 
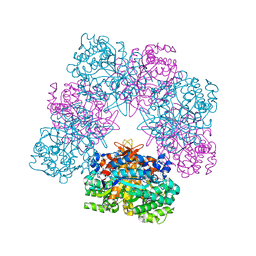 | | Structure of beta-glucosidase A from Paenibacillus polymyxa complexed with a monovalent inhibitor | | Descriptor: | (2~{S},3~{S},4~{R})-2-[[4-[4-[2-[2-(2-azanylidenehydrazinyl)ethoxy]ethoxy]phenyl]-1,2,3-triazol-1-yl]methyl]pyrrolidine-3,4-diol, Beta-glucosidase A | | Authors: | Jimenez-Ortega, E, Sanz-Aparicio, J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-07-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structural basis of the inhibition of GH1 beta-glucosidases by multivalent pyrrolidine iminosugars.
Bioorg.Chem., 89, 2019
|
|
5MI5
 
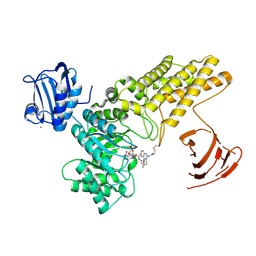 | | BtGH84 mutant with covalent modification by MA3 in complex with PUGNAc | | Descriptor: | CALCIUM ION, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE, O-GlcNAcase BT_4395, ... | | Authors: | Darby, J.F, Davies, G.J, Hubbard, R.E. | | Deposit date: | 2016-11-27 | | Release date: | 2017-11-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Increase of enzyme activity through specific covalent modification with fragments.
Chem Sci, 8, 2017
|
|
6REG
 
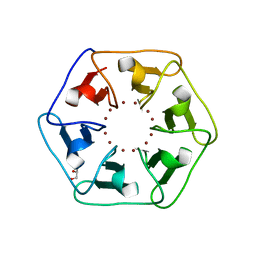 | | Crystal structure of Pizza6-S with Zn2+ | | Descriptor: | GLYCEROL, Pizza6-S, ZINC ION | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6REK
 
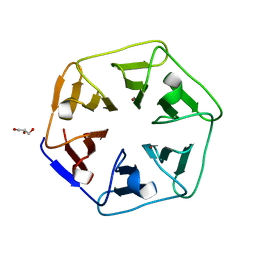 | | Crystal structure of Pizza6-SH with Cu2+ | | Descriptor: | COPPER (II) ION, GLYCEROL, Pizza6-SH | | Authors: | Noguchi, H, Clarke, D.E, Gryspeerdt, J.L, Feyter, S.D, Voet, A.R.D. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Artificial beta-propeller protein-based hydrolases.
Chem.Commun.(Camb.), 55, 2019
|
|
6R7D
 
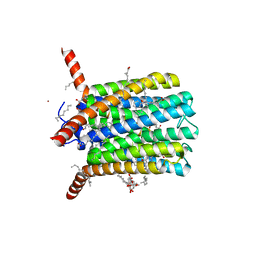 | | Crystal structure of LTC4S in complex with AZ13690257 | | Descriptor: | (1~{S},2~{S})-2-[5-[cyclopropylmethyl(naphthalen-1-yl)amino]-4-methoxy-pyrimidin-2-yl]carbonylcyclopropane-1-carboxylic acid, DODECYL-BETA-D-MALTOSIDE, Leukotriene C4 synthase, ... | | Authors: | Kack, H, Ek, M. | | Deposit date: | 2019-03-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of the Oral Leukotriene C4 Synthase Inhibitor (1S,2S)-2-({5-[(5-Chloro-2,4-difluorophenyl)(2-fluoro-2-methylpropyl)amino]-3-methoxypyrazin-2-yl}carbonyl)cyclopropanecarboxylic Acid (AZD9898) as a New Treatment for Asthma.
J.Med.Chem., 62, 2019
|
|
5MKU
 
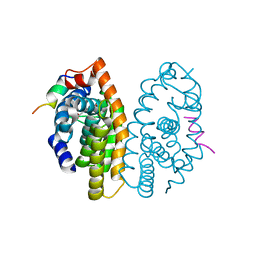 | | Crystal structure of the Retinoid X Receptor alpha in complex with synthetic honokiol derivative 4 and a fragment of the TIF2 co-activator. | | Descriptor: | (~{E})-3-[4-oxidanyl-3-(3-propan-2-ylphenyl)phenyl]prop-2-enoic acid, HIS-LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN-ASP-SER, Retinoic acid receptor RXR-alpha | | Authors: | Andrei, S.A, Brunsveld, L, Scheepstra, M, Ottmann, C. | | Deposit date: | 2016-12-05 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Ligand Dependent Switch from RXR Homo- to RXR-NURR1 Heterodimerization.
ACS Chem Neurosci, 8, 2017
|
|
6RBS
 
 | |
6RBZ
 
 | |
6R7S
 
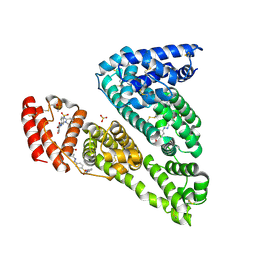 | | Human Serum Albumin, complexed with Sulfasalazine | | Descriptor: | 2-HYDROXY-(5-([4-(2-PYRIDINYLAMINO)SULFONYL]PHENYL)AZO)BENZOIC ACID, DIMETHYL SULFOXIDE, SULFATE ION, ... | | Authors: | Schreuder, H.A, Liesum, A. | | Deposit date: | 2019-03-29 | | Release date: | 2020-04-08 | | Last modified: | 2020-05-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Resolving Binding Events on the Multifunctional Human Serum Albumin.
Chemmedchem, 15, 2020
|
|
6RC9
 
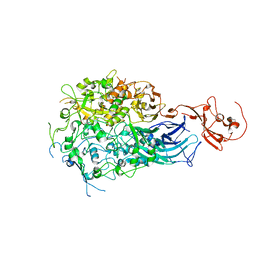 | |
5MMD
 
 | | TMB-1. Structural insights into TMB-1 and the role of residue 119 and 228 in substrate and inhibitor binding | | Descriptor: | CHLORIDE ION, Metallo-beta-lactamase 1, ZINC ION | | Authors: | Skagseth, S, Christopeit, T, Akhter, S, Bayer, A, Samuelsen, O, Leiros, H.-K.S. | | Deposit date: | 2016-12-09 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights into TMB-1 and the Role of Residues 119 and 228 in Substrate and Inhibitor Binding.
Antimicrob. Agents Chemother., 61, 2017
|
|
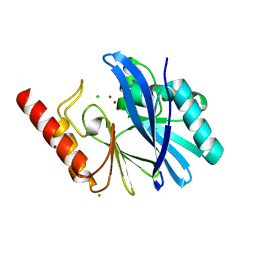
5MQS
 
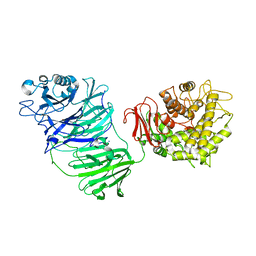 | | Sialidase BT_1020 | | Descriptor: | Beta-L-arabinobiosidase, CALCIUM ION, SODIUM ION, ... | | Authors: | Basle, A, Ndeh, D, Rogowski, A, Cartmell, A, Luis, A.S, Venditto, I, Labourel, A, Gilbert, H.J. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Complex pectin metabolism by gut bacteria reveals novel catalytic functions.
Nature, 544, 2017
|
|
6R9H
 
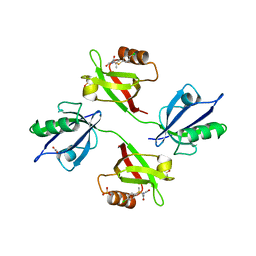 | | Crystal structure of the PDZ tandem of syntenin in complex with fragment C58 | | Descriptor: | (2~{S})-2-[2-(4-chlorophenyl)sulfanylethanoylamino]-3-methyl-butanoic acid, ACETATE ION, PHOSPHATE ION, ... | | Authors: | Feracci, M, Barral, K. | | Deposit date: | 2019-04-03 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pharmacological inhibition of syntenin PDZ2 domain impairs breast cancer cell activities and exosome loadifing with syndecan and EpCAM cargo.
J Extracell Vesicles, 10, 2020
|
|
5MR4
 
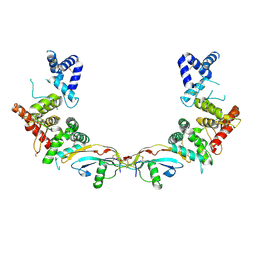 | | Ligand-receptor complex. | | Descriptor: | DI(HYDROXYETHYL)ETHER, FORMIC ACID, GDNF family receptor alpha-2, ... | | Authors: | Sandmark, J, Oster, L, Aagaard, A, Roth, R.G, Dahl, G. | | Deposit date: | 2016-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and biophysical characterization of the human full-length neurturin-GFRa2 complex: A role for heparan sulfate in signaling.
J. Biol. Chem., 293, 2018
|
|
5MO9
 
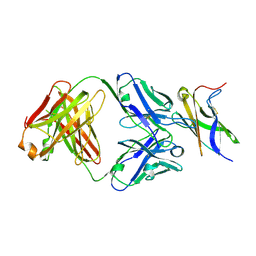 | |
5MRL
 
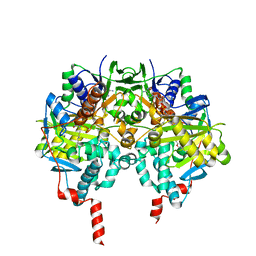 | | Crystal structure of human monoamine oxidase B (MAO B) in complex with N(Furan2ylmethyl)Nmethylprop2yn1amine (F2MPA) | | Descriptor: | (~{E})-~{N}-(furan-2-ylmethyl)-~{N}-methyl-prop-1-en-1-amine, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, De Deurwaerdere, P, Corne, R, Leone, C, Valeri, A, Valoti, M, Ramsay, R.R, Fall, Y, Marco-Contelles, J. | | Deposit date: | 2016-12-23 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Comparative Analysis of the Neurochemical Profile and MAO Inhibition Properties of N-(Furan-2-ylmethyl)-N-methylprop-2-yn-1-amine.
ACS Chem Neurosci, 8, 2017
|
|
5MOI
 
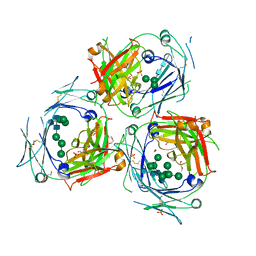 | | Crystal structure of human IgE-Fc epsilon 3-4 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ig epsilon chain C region, ... | | Authors: | Dore, K.A, Davies, A.M, Drinkwater, N, Beavil, A.J, McDonnell, J.M, Sutton, B.J. | | Deposit date: | 2016-12-14 | | Release date: | 2018-01-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Thermal sensitivity and flexibility of the C epsilon 3 domains in immunoglobulin E.
Biochim. Biophys. Acta, 1865, 2017
|
|
