2MZ1
 
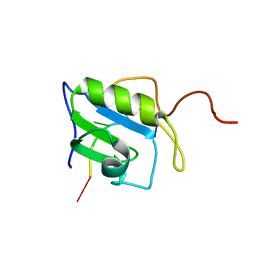 | | Solution structure of hnRNP C RRM in complex with 5'-UUUUC-3' RNA | | Descriptor: | 5'-R(*UP*UP*UP*UP*C)-3', Heterogeneous nuclear ribonucleoproteins C1/C2 | | Authors: | Cienikova, Z, Damberger, F.F, Hall, J, Allain, F.H.-T, Maris, C. | | Deposit date: | 2015-02-05 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and mechanistic insights into poly(uridine) tract recognition by the hnRNP C RNA recognition motif.
J.Am.Chem.Soc., 136, 2014
|
|
5MQF
 
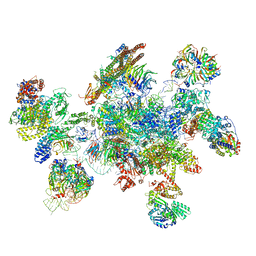 | | Cryo-EM structure of a human spliceosome activated for step 2 of splicing (C* complex) | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, ATP-dependent RNA helicase DHX8, Cell division cycle 5-like protein, ... | | Authors: | Bertram, K, Hartmuth, K, Kastner, B. | | Deposit date: | 2016-12-20 | | Release date: | 2017-03-22 | | Last modified: | 2018-11-21 | | Method: | ELECTRON MICROSCOPY (5.9 Å) | | Cite: | Cryo-EM structure of a human spliceosome activated for step 2 of splicing.
Nature, 542, 2017
|
|
5LQF
 
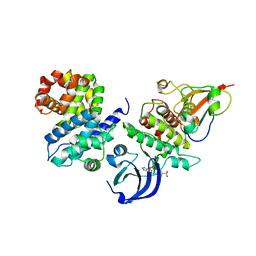 | | CDK1/CyclinB1/CKS2 in complex with NU6102 | | Descriptor: | Cyclin-dependent kinase 1, Cyclin-dependent kinases regulatory subunit 2, G2/mitotic-specific cyclin-B1, ... | | Authors: | Coxon, C.R, Anscombe, E, Harnor, S.J, Martin, M.P, Carbain, B.J, Hardcastle, I.R, Harlow, L.K, Korolchuk, S, Matheson, C.J, Noble, M.E, Newell, D.R, Turner, D.M, Sivaprakasam, M, Wang, L.Z, Wong, C, Golding, B.T, Griffin, R.J, Endicott, J.A, Cano, C. | | Deposit date: | 2016-08-17 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Cyclin-Dependent Kinase (CDK) Inhibitors: Structure-Activity Relationships and Insights into the CDK-2 Selectivity of 6-Substituted 2-Arylaminopurines.
J. Med. Chem., 60, 2017
|
|
3JAH
 
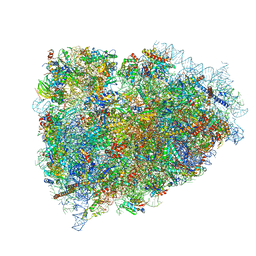 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UAG stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
3IYG
 
 | | Ca model of bovine TRiC/CCT derived from a 4.0 Angstrom cryo-EM map | | Descriptor: | T-complex protein 1 subunit, T-complex protein 1 subunit alpha, T-complex protein 1 subunit beta, ... | | Authors: | Cong, Y, Baker, M.L, Ludtke, S.J, Frydman, J, Chiu, W. | | Deposit date: | 2009-11-28 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | 4.0-A resolution cryo-EM structure of the mammalian chaperonin TRiC/CCT reveals its unique subunit arrangement.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5MPG
 
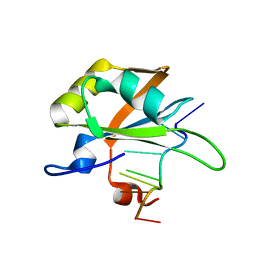 | |
3JAG
 
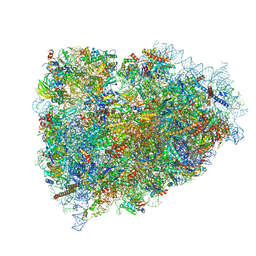 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UAA stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
5O9Z
 
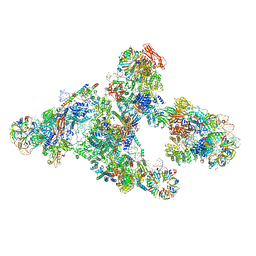 | |
3JAI
 
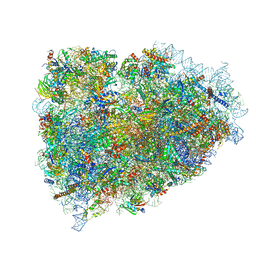 | | Structure of a mammalian ribosomal termination complex with ABCE1, eRF1(AAQ), and the UGA stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Brown, A, Shao, S, Murray, J, Hegde, R.S, Ramakrishnan, V. | | Deposit date: | 2015-06-10 | | Release date: | 2015-08-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for stop codon recognition in eukaryotes.
Nature, 524, 2015
|
|
8U17
 
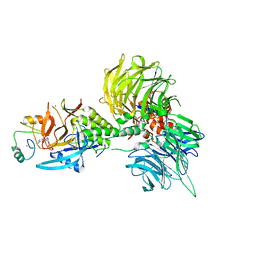 | | The ternary complex structure of DDB1-CRBN-SALL4(ZF1,2)-long bound to Pomalidomide | | Descriptor: | DNA damage-binding protein 1, Protein cereblon, S-Pomalidomide, ... | | Authors: | Clifton, M.C, Ma, X, Ornelas, E. | | Deposit date: | 2023-08-30 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and biophysical comparisons of the pomalidomide- and CC-220-induced interactions of SALL4 with cereblon.
Sci Rep, 13, 2023
|
|
8U14
 
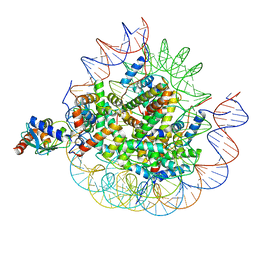 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A lysine 15 in complex with RNF168-UbcH5c (class 2) | | Descriptor: | DNA (146-MER), DNA (147-MER), E3 ubiquitin-protein ligase RNF168, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-30 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TGP
 
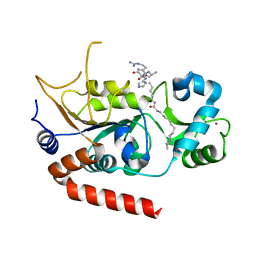 | |
8TXX
 
 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 3) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TXV
 
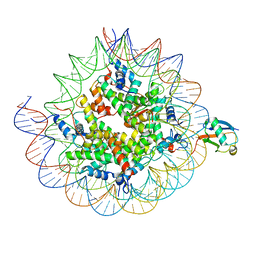 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 1) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8U13
 
 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A lysine 15 in complex with RNF168-UbcH5c (class 1) | | Descriptor: | DNA (146-MER), DNA (147-MER), E3 ubiquitin-protein ligase RNF168, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-30 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TXW
 
 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 2) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
2RO1
 
 | | NMR Solution Structures of Human KAP1 PHD finger-bromodomain | | Descriptor: | Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Zeng, L, Yap, K.L, Ivanov, A.V, Wang, X, Mujtaba, S, Plotnikova, O, Rauscher, F.J. | | Deposit date: | 2008-03-04 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into human KAP1 PHD finger-bromodomain and its role in gene silencing
Nat.Struct.Mol.Biol., 15, 2008
|
|
8UPF
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168-UbcH5c | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-10-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
5WWE
 
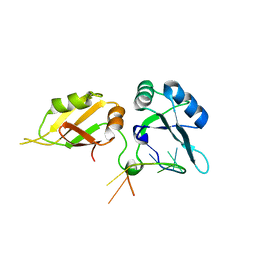 | | Crystal structure of hnRNPA2B1 in complex with RNA | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*GP*GP*GP*AP*CP*UP*AP*GP*A)-3') | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
5WWG
 
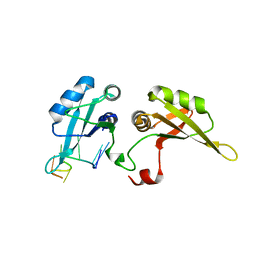 | | Crystal structure of hnRNPA2B1 in complex with AAGGACUUGC | | Descriptor: | Heterogeneous nuclear ribonucleoproteins A2/B1, RNA (5'-R(*AP*AP*GP*GP*AP*CP*UP*U)-3') | | Authors: | Wu, B.X, Su, S.C, Ma, J.B. | | Deposit date: | 2017-01-01 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1.
Nat Commun, 9, 2018
|
|
2UYY
 
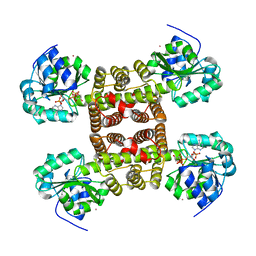 | | Structure of the cytokine-like nuclear factor n-pac | | Descriptor: | N-PAC PROTEIN, POTASSIUM ION, [(2R,3R,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3-HYDROXY-4-(PHOSPHONOOXY)TETRAHYDROFURAN-2-YL]METHYL [(2R,3S,4S)-3,4-DIHYDROXYTETRAHYDROFURAN-2-YL]METHYL DIHYDROGEN DIPHOSPHATE | | Authors: | Tickle, J, Pilka, E.S, Bunkoczi, G, Berridge, G, Smee, C, Kavanagh, K.L, Hozjan, V, Niesen, F.H, Papagrigoriou, E, Pike, A.C.W, Turnbull, A, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, von Delft, F, Oppermann, U. | | Deposit date: | 2007-04-20 | | Release date: | 2007-07-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of the Cytokine-Like Nuclear Factor N-Pac
To be Published
|
|
2XB2
 
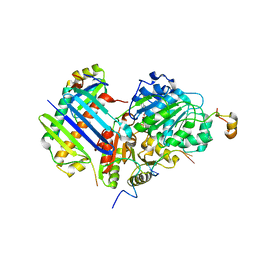 | | Crystal structure of the core Mago-Y14-eIF4AIII-Barentsz-UPF3b assembly shows how the EJC is bridged to the NMD machinery | | Descriptor: | EUKARYOTIC INITIATION FACTOR 4A-III, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Buchwald, G, Ebert, J, Basquin, C, Sauliere, J, Jayachandran, U, Bono, F, Le Hir, H, Conti, E. | | Deposit date: | 2010-04-03 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Insights Into the Recruitment of the Nmd Machinery from the Crystal Structure of a Core Ejc-Upf3B Complex.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6H25
 
 | | Human nuclear RNA exosome EXO-10-MPP6 complex | | Descriptor: | Exosome complex component CSL4, Exosome complex component MTR3, Exosome complex component RRP4, ... | | Authors: | Gerlach, P, Schuller, J.M, Falk, S, Basquin, J, Conti, E. | | Deposit date: | 2018-07-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Distinct and evolutionary conserved structural features of the human nuclear exosome complex.
Elife, 7, 2018
|
|
6HCM
 
 | | Structure of the rabbit collided di-ribosome (stalled monosome) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-15 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
6HCF
 
 | | Structure of the rabbit 80S ribosome stalled on globin mRNA at the stop codon | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Juszkiewicz, S, Chandrasekaran, V, Lin, Z, Kraatz, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2018-08-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ZNF598 Is a Quality Control Sensor of Collided Ribosomes.
Mol. Cell, 72, 2018
|
|
