5GYZ
 
 | | luciferase AMP/7-cy-L complex | | Descriptor: | (4S)-2-[6-(azepan-1-yl)-1,3-benzothiazol-2-yl]-4,5-dihydro-1,3-thiazole-4-carboxylic acid, ADENOSINE MONOPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Su, J, Wang, F. | | Deposit date: | 2016-09-26 | | Release date: | 2017-09-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of luciferase with AMP/7-cy-L at 2.4 Angstroms resolution
To Be Published
|
|
3GXO
 
 | | Structure of the Mitomycin 7-O-methyltransferase MmcR with bound Mitomycin A | | Descriptor: | CALCIUM ION, MmcR, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Singh, S, Chang, A, Bingman, C.A, Phillips Jr, G.N, Thorson, J.S. | | Deposit date: | 2009-04-02 | | Release date: | 2010-04-21 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural characterization of the mitomycin 7-O-methyltransferase.
Proteins, 79, 2011
|
|
3BUS
 
 | | Crystal Structure of RebM | | Descriptor: | Methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | McCoy, J.G, Singh, S, Bingman, C.A, Thorson, J.S, Phillips Jr, G.N. | | Deposit date: | 2008-01-03 | | Release date: | 2008-03-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and mechanism of the rebeccamycin sugar 4'-O-methyltransferase RebM.
J.Biol.Chem., 283, 2008
|
|
4OKO
 
 | | Crystal structure of Francisella tularensis REP34 (Rapid Encystment Phenotype Protein 34 KDa) | | Descriptor: | ACETATE ION, Rapid Encystment Phenotype Protein 34 KDa, ZINC ION | | Authors: | Feld, G.K, Segelke, B.W, Corzett, M.H, Hunter, M.S, Frank, M, Rasley, A. | | Deposit date: | 2014-01-22 | | Release date: | 2014-09-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.053 Å) | | Cite: | Structure and Function of REP34 Implicates Carboxypeptidase Activity in Francisella tularensis Host Cell Invasion.
J.Biol.Chem., 289, 2014
|
|
4H3Y
 
 | |
1TW2
 
 | | Crystal structure of Carminomycin-4-O-methyltransferase (DnrK) in complex with S-adenosyl-L-homocystein (SAH) and 4-methoxy-e-rhodomycin T (M-ET) | | Descriptor: | Carminomycin 4-O-methyltransferase, METHYL (4R)-2-ETHYL-2,5,12-TRIHYDROXY-7-METHOXY-6,11-DIOXO-4-{[2,3,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-RIBO-HEXOPYRANOSYL]OXY}-1H,2H,3H,4H,6H,11H-TETRACENE-1-CARBOXYLATE, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Jansson, A, Koskiniemi, H, Mantsala, P, Niemi, J, Schneider, G, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-30 | | Release date: | 2004-09-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a ternary complex of DnrK, a methyltransferase in daunorubicin biosynthesis, with bound products
J.Biol.Chem., 279, 2004
|
|
2OPS
 
 | | Crystal Structure of Y188C Mutant HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Short, S.A, Chan, J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
2OPQ
 
 | | Crystal Structure of L100I Mutant HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Short, S.A, Chan, J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
1D2C
 
 | | METHYLTRANSFERASE | | Descriptor: | PROTEIN (GLYCINE N-METHYLTRANSFERASE) | | Authors: | Huang, Y, Takusagawa, F. | | Deposit date: | 1999-09-23 | | Release date: | 1999-10-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanisms for auto-inhibition and forced product release in glycine N-methyltransferase: crystal structures of wild-type, mutant R175K and S-adenosylhomocysteine-bound R175K enzymes.
J.Mol.Biol., 298, 2000
|
|
6ZK0
 
 | | 1.47A human IMPase with ebselen | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Inositol monophosphatase 1, ... | | Authors: | Bax, B.D, Fenn, G.D. | | Deposit date: | 2020-06-29 | | Release date: | 2020-09-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallization and structure of ebselen bound to Cys141 of human inositol monophosphatase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5GS7
 
 | |
2OPP
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Chan, S.J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
3O7B
 
 | |
1NC1
 
 | | Crystal structure of E. coli MTA/AdoHcy nucleosidase complexed with 5'-methylthiotubercidin (MTH) | | Descriptor: | 2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-METHYLSULFANYLMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, MTA/SAH nucleosidase | | Authors: | Lee, J.E, Cornell, K.A, Riscoe, M.K, Howell, P.L. | | Deposit date: | 2002-12-04 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Escherichia coli 5'-methylthioadenosine/ S-adenosylhomocysteine nucleosidase inhibitor complexes provide insight into the conformational changes required for substrate binding and catalysis.
J.Biol.Chem., 278, 2003
|
|
1QJE
 
 | | Isopenicillin N synthase from Aspergillus nidulans (IP1 - Fe complex) | | Descriptor: | FE (II) ION, ISOPENICILLIN N, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Burzlaff, N.I, Clifton, I.J, Rutledge, P.J, Roach, P.L, Adlington, R.M, Baldwin, J.E. | | Deposit date: | 1999-06-23 | | Release date: | 2000-06-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Reaction Cycle of Isopenicillin N Synthase Observed by X-Ray Diffraction
Nature, 401, 1999
|
|
8DOM
 
 | |
5HU4
 
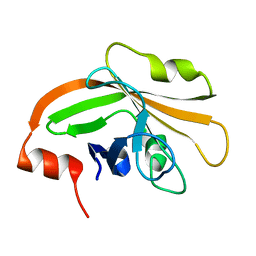 | | Cystal structure of listeria monocytogenes sortase A | | Descriptor: | Cysteine protease | | Authors: | Li, H. | | Deposit date: | 2016-01-27 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Inhibition of sortase A by chalcone prevents Listeria monocytogenes infection.
Biochem. Pharmacol., 106, 2016
|
|
7SU8
 
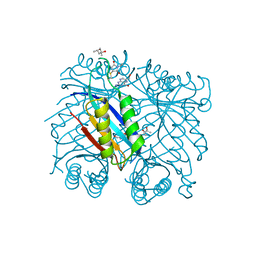 | |
2K0R
 
 | | Solution structure of the C103S mutant of the N-terminal Domain of DsbD from Neisseria meningitidis | | Descriptor: | Thiol:disulfide interchange protein dsbD | | Authors: | Quinternet, M, Selme, L, Tsan, P, Beaufils, C, Jacob, C, Boschi-Muller, S, Averlant-Petit, M, Branlant, G, Cung, M. | | Deposit date: | 2008-02-13 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the cysteine 103 to serine mutant of the N-terminal domain of DsbD from Neisseria meningitidis.
Biochemistry, 47, 2008
|
|
3OFK
 
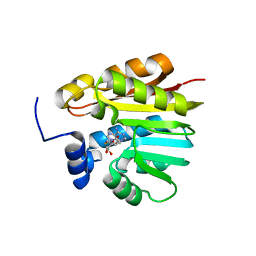 | | Crystal structure of N-methyltransferase NodS from Bradyrhizobium japonicum WM9 in complex with S-adenosyl-l-homocysteine (SAH) | | Descriptor: | Nodulation protein S, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cakici, O, Sikorski, M, Stepkowski, T, Bujacz, G, Jaskolski, M. | | Deposit date: | 2010-08-15 | | Release date: | 2010-10-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of NodS N-Methyltransferase from Bradyrhizobium japonicum in Ligand-Free Form and as SAH Complex.
J.Mol.Biol., 404, 2010
|
|
2M77
 
 | | [Asp2]RTD-1 | | Descriptor: | [Asp2]RTD-1 | | Authors: | Conibear, A.C, Bochen, A, Rosengren, K, Kessler, H, Craik, D.J. | | Deposit date: | 2013-04-18 | | Release date: | 2014-02-05 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | The Cyclic Cystine Ladder of Theta-Defensins as a Stable, Bifunctional Scaffold: A Proof-of-Concept Study Using the Integrin-Binding RGD Motif
Chembiochem, 15, 2014
|
|
8DSK
 
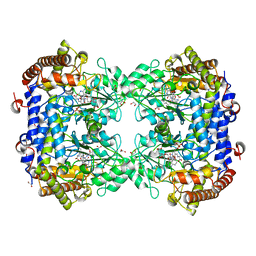 | | Structure of the N358Y variant of serine hydroxymethyltransferase 8 in complex with PLP, glycine, and formyl tetrahydrofolate | | Descriptor: | 1,2-ETHANEDIOL, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, ... | | Authors: | Korasick, D.A, Beamer, L.J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-10-18 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural and functional analysis of two SHMT8 variants associated with soybean cyst nematode resistance.
Febs J., 291, 2024
|
|
4G56
 
 | | Crystal Structure of full length PRMT5/MEP50 complexes from Xenopus laevis | | Descriptor: | Hsl7 protein, MGC81050 protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Ho, M, Wilczek, C, Bonanno, J, Shechter, D, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structure of the arginine methyltransferase PRMT5-MEP50 reveals a mechanism for substrate specificity
Plos One, 8, 2013
|
|
2OPR
 
 | | Crystal Structure of K101E Mutant HIV-1 Reverse Transcriptase in Complex with GW420867X. | | Descriptor: | ISOPROPYL (2S)-2-ETHYL-7-FLUORO-3-OXO-3,4-DIHYDROQUINOXALINE-1(2H)-CARBOXYLATE, Reverse transcriptase/ribonuclease H, p51 RT | | Authors: | Ren, J, Nichols, C.E, Chamberlain, P.P, Weaver, K.L, Short, S.A, Chan, J.H, Kleim, J, Stammers, D.K. | | Deposit date: | 2007-01-30 | | Release date: | 2007-05-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Relationship of Potency and Resilience to Drug Resistant Mutations for GW420867X Revealed by Crystal Structures of Inhibitor Complexes for Wild-Type, Leu100Ile, Lys101Glu, and Tyr188Cys Mutant HIV-1 Reverse Transcriptases.
J.Med.Chem., 50, 2007
|
|
4G95
 
 | | hDHFR-OAG binary complex | | Descriptor: | 6-{[(2,5-dichlorophenyl)amino]methyl}pyrido[2,3-d]pyrimidine-2,4-diamine, Dihydrofolate reductase, SULFATE ION | | Authors: | Cody, V, Pace, J, Queener, S, Adair, O, Gangjee, A. | | Deposit date: | 2012-07-23 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Kinetic and structural analysis for potent antifolate inhibition of Pneumocystis jirovecii, Pneumocystis carinii, and human dihydrofolate reductases and their active-site variants.
Antimicrob.Agents Chemother., 57, 2013
|
|
