2PVH
 
 | | Structure-Based Design of Pyrazolo[1,5-a][1,3,5]triazine Derivatives as Potent Inhibitors of Protein Kinase CK2 | | Descriptor: | Casein kinase II subunit alpha, N,N'-DIPHENYLPYRAZOLO[1,5-A][1,3,5]TRIAZINE-2,4-DIAMINE | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Almassy, R, Lu, J, Averill, A, Yager, K.M, Chu, S. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
4IVQ
 
 | | Crystal structure of thymidine kinase from Herpes simplex virus type 1 in complex with IN43/5 | | Descriptor: | 6-[3-hydroxy-2-(hydroxymethyl)prop-1-en-1-yl]-4-methoxy-1,5-dimethylpyrimidin-2(1H)-one, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Novakovic, I, Westermaier, Y, Perozzo, R, Raic-Malic, S, Scapozza, L. | | Deposit date: | 2013-01-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New thymine derivatives as HSV1-TK ligand for the development of PET imaging tracer
To be Published
|
|
2PVM
 
 | | Structure-Based Design of Pyrazolo[1,5-a][1,3,5]triazine Derivatives as Potent Inhibitors of Protein Kinase CK2 | | Descriptor: | 4-(2-(1H-IMIDAZOL-4-YL)ETHYLAMINO)-2-(PHENYLAMINO)PYRAZOLO[1,5-A][1,3,5]TRIAZINE-8-CARBONITRILE, Casein kinase II subunit alpha | | Authors: | Nie, Z, Perretta, C, Erickson, P, Margosiak, S, Almassy, R, Lu, J, Averill, A, Yager, K.M, Chu, S. | | Deposit date: | 2007-05-09 | | Release date: | 2008-05-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis, and study of pyrazolo[1,5-a][1,3,5]triazine derivatives as potent inhibitors of protein kinase CK2.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3QLH
 
 | | HIV-1 Reverse Transcriptase in Complex with Manicol at the RNase H Active Site and TMC278 (Rilpivirine) at the NNRTI Binding Pocket | | Descriptor: | (2S)-5,7-dihydroxy-9-methyl-2-(prop-1-en-2-yl)-1,2,3,4-tetrahydro-6H-benzo[7]annulen-6-one, 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Himmel, D.M, Wojtak, K, Bauman, J.D, Arnold, E. | | Deposit date: | 2011-02-02 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthesis, activity, and structural analysis of novel alpha-hydroxytropolone inhibitors of human immunodeficiency virus reverse transcriptase-associated ribonuclease H.
J.Med.Chem., 54, 2011
|
|
3BWH
 
 | | Atomic resolution structure of cucurmosin, a novel type 1 RIP from the sarcocarp of Cucurbita moschata | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, beta-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)][alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, L. | | Deposit date: | 2008-01-09 | | Release date: | 2008-10-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Atomic resolution structure of cucurmosin, a novel type 1 ribosome-inactivating protein from the sarcocarp of Cucurbita moschata.
J.Struct.Biol., 164, 2008
|
|
1DOJ
 
 | | Crystal structure of human alpha-thrombin*RWJ-51438 complex at 1.7 A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN, HIRUGEN, ... | | Authors: | Recacha, R, Costanzo, M.J, Maryanoff, B.E, Carson, M, DeLucas, L, Chattopadhyay, D. | | Deposit date: | 1999-12-21 | | Release date: | 2000-11-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of human alpha-thrombin complexed with RWJ-51438 at 1.7 A: unusual perturbation of the 60A-60I insertion loop.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
3R6B
 
 | | Crystal Structure of Thrombospondin-1 TSR Domains 2 and 3 | | Descriptor: | 1,2-ETHANEDIOL, Thrombospondin-1 | | Authors: | Page, R.C, Klenotic, P.A, Misra, S, Silverstein, R.L. | | Deposit date: | 2011-03-21 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Expression, purification and structural characterization of functionally replete thrombospondin-1 type 1 repeats in a bacterial expression system.
Protein Expr.Purif., 80, 2011
|
|
4KKO
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 4-((4-methoxy-6-(2-morpholinoethoxy)-1,3,5-triazin-2-yl)amino)-2-((3-methylbut-2-en-1-yl)oxy)benzonitrile (JLJ513), a non-nucleoside inhibitor | | Descriptor: | 4-({4-methoxy-6-[2-(morpholin-4-yl)ethoxy]-1,3,5-triazin-2-yl}amino)-2-(3-methylbutoxy)benzonitrile, HIV-1 reverse transcriptase, p51 subunit, ... | | Authors: | Frey, K.M, Anderson, K.S. | | Deposit date: | 2013-05-06 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Extension into the entrance channel of HIV-1 reverse transcriptase-Crystallography and enhanced solubility.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1TW5
 
 | | beta1,4-galactosyltransferase mutant M344H-Gal-T1 in complex with Chitobiose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ramakrishnan, B, Boeggeman, E, Qasba, P.K. | | Deposit date: | 2004-06-30 | | Release date: | 2004-12-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Effect of the Met344His mutation on the conformational dynamics of bovine beta-1,4-galactosyltransferase: crystal structure of the Met344His mutant in complex with chitobiose
Biochemistry, 43, 2004
|
|
1XL0
 
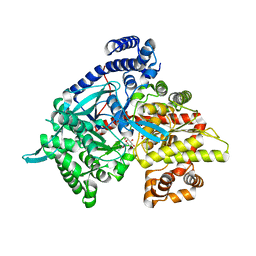 | | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1,3,4-oxadiazole,-benzothiazole, and-benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site. | | Descriptor: | (1R)-1,5-anhydro-1-(5-methyl-1,3,4-oxadiazol-2-yl)-D-glucitol, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Kosmopoulou, M.N, Tiraidis, C, Kardakaris, R, Bischler, N, Leonidas, D.D, Hadady, Z, Somsak, L, Docsa, T, Gergely, P, Oikonomakos, N.G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1, 3, 4-oxadiazole, -benzothiazole, and -benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site
Protein Sci., 14, 2005
|
|
1XL1
 
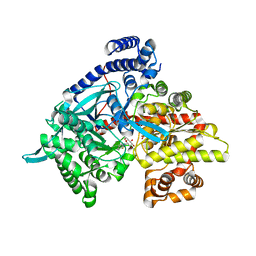 | | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1,3,4-oxadiazole,-benzothiazole, and-benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site. | | Descriptor: | (1R)-1,5-anhydro-1-(5-methyl-1,3-benzothiazol-2-yl)-D-glucitol, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Kosmopoulou, M.N, Tiraidis, C, Kardakaris, R, Bischler, N, Leonidas, D.D, Hadady, Z, Somsak, L, Docsa, T, Gergely, P, Oikonomakos, N.G. | | Deposit date: | 2004-09-30 | | Release date: | 2005-03-15 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and crystallographic studies on 2-(beta-D-glucopyranosyl)-5-methyl-1, 3, 4-oxadiazole, -benzothiazole, and -benzimidazole, inhibitors of muscle glycogen phosphorylase b. Evidence for a new binding site
Protein Sci., 14, 2005
|
|
3OXC
 
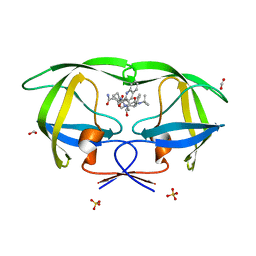 | | Wild Type HIV-1 Protease with Antiviral Drug Saquinavir | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, FORMIC ACID, Protease, ... | | Authors: | Kovalevsky, A.Y, Wang, Y.-F, Tie, Y, Weber, I.T. | | Deposit date: | 2010-09-21 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir
Proteins, 67, 2007
|
|
3D20
 
 | | Crystal structure of HIV-1 mutant I54V and inhibitor DARUNAVIA | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, HIV-1 Protease, ... | | Authors: | Liu, F, Kovalesky, A.Y, Tie, Y, Ghosh, A.K, Harrison, R.W, Weber, I.T. | | Deposit date: | 2008-05-07 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
3OXW
 
 | | Crystal Structure of HIV-1 I50V, A71V Protease in Complex with the Protease Inhibitor Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, 1,2-ETHANEDIOL, ACETATE ION, ... | | Authors: | Mittal, S, Bandaranayake, R.M, Schiffer, C.A. | | Deposit date: | 2010-09-22 | | Release date: | 2011-09-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and thermodynamic basis of amprenavir/darunavir and atazanavir resistance in HIV-1 protease with mutations at residue 50.
J.Virol., 87, 2013
|
|
3D1Y
 
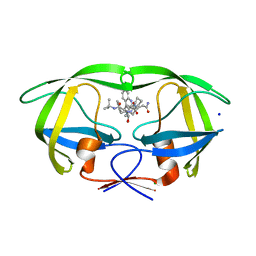 | | Crystal structure of HIV-1 mutant I54V and inhibitor SAQUINA | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, CHLORIDE ION, HIV-1 Protease, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-05-06 | | Release date: | 2008-05-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
J.Mol.Biol., 381, 2008
|
|
3SO9
 
 | | Darunavir in Complex with a Human Immunodeficiency Virus Type 1 Protease Variant | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 protease | | Authors: | Wang, Y, Liu, Z, Brunzelle, S.J, Kovari, L.C, Kovari, I.A. | | Deposit date: | 2011-06-30 | | Release date: | 2011-10-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | The higher barrier of darunavir and tipranavir resistance for HIV-1 protease.
Biochem.Biophys.Res.Commun., 412, 2011
|
|
1BOT
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX BETWEEN ESCHERICHIA COLI GLYCEROL KINASE AND THE ALLOSTERIC REGULATOR FRUCTOSE 1,6-BISPHOSPHATE. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, PROTEIN (GLYCEROL KINASE) | | Authors: | Ormo, M, Bystrom, C.E, Remington, S.J. | | Deposit date: | 1998-08-05 | | Release date: | 1999-01-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of a complex of Escherichia coli glycerol kinase and an allosteric effector fructose 1,6-bisphosphate.
Biochemistry, 37, 1998
|
|
3SFU
 
 | | crystal structure of murine norovirus RNA dependent RNA polymerase in complex with ribavirin | | Descriptor: | 1-(beta-D-ribofuranosyl)-1H-1,2,4-triazole-3-carboxamide, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Kim, K.H, Alam, I. | | Deposit date: | 2011-06-14 | | Release date: | 2012-05-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Crystal structures of murine norovirus-1 RNA-dependent RNA polymerase in complex with 2-thiouridine or ribavirin.
Virology, 426, 2012
|
|
2HB3
 
 | | Wild-type HIV-1 Protease in complex with potent inhibitor GRL06579 | | Descriptor: | (3AS,5R,6AR)-HEXAHYDRO-2H-CYCLOPENTA[B]FURAN-5-YL (2S,3S)-3-HYDROXY-4-(4-(HYDROXYMETHYL)-N-ISOBUTYLPHENYLSULFONAMIDO)-1-PHENYLBUTAN-2-YLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Design of Novel HIV-1 Protease Inhibitors To Combat Drug Resistance.
J.Med.Chem., 49, 2006
|
|
1DIF
 
 | | HIV-1 PROTEASE IN COMPLEX WITH A DIFLUOROKETONE CONTAINING INHIBITOR A79285 | | Descriptor: | BETA-MERCAPTOETHANOL, HIV-1 PROTEASE, N-{1-BENZYL-2,2-DIFLUORO-3,3-DIHYDROXY-4-[3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRYLAMINO]-5-PHENYL-PENTYL}-3-METHYL-2-(3-METHYL-3-PYRIDIN-2-YLMETHYL-UREIDO)-BUTYRAMIDE | | Authors: | Silva, A.M, Cachau, R.E, Sham, H.L, Erickson, J.W. | | Deposit date: | 1995-10-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition and catalytic mechanism of HIV-1 aspartic protease.
J.Mol.Biol., 255, 1996
|
|
1EQC
 
 | | EXO-B-(1,3)-GLUCANASE FROM CANDIDA ALBICANS IN COMPLEX WITH CASTANOSPERMINE AT 1.85 A | | Descriptor: | CASTANOSPERMINE, EXO-(B)-(1,3)-GLUCANASE | | Authors: | Cutfield, S.M, Davies, G.J, Murshudov, G, Anderson, B.F, Moody, P.C.E, Sullivan, P.A, Cutfield, J.F. | | Deposit date: | 2000-04-03 | | Release date: | 2000-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of the exo-beta-(1,3)-glucanase from Candida albicans in native and bound forms: relationship between a pocket and groove in family 5 glycosyl hydrolases.
J.Mol.Biol., 294, 1999
|
|
1C6X
 
 | |
1C6Y
 
 | |
1RPK
 
 | | Crystal structure of barley alpha-amylase isozyme 1 (amy1) in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase type 1 isozyme, ... | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2003-12-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Binding to Barley {alpha}-Amylase 1
J.Biol.Chem., 280, 2005
|
|
1AXA
 
 | | ACTIVE-SITE MOBILITY IN HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 PROTEASE AS DEMONSTRATED BY CRYSTAL STRUCTURE OF A28S MUTANT | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Hartsuck, J.A, Foundling, S, Ermolieff, J, Tang, J. | | Deposit date: | 1997-10-13 | | Release date: | 1998-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active-site mobility in human immunodeficiency virus, type 1, protease as demonstrated by crystal structure of A28S mutant.
Protein Sci., 7, 1998
|
|
