3F0T
 
 | | Crystal structure of thymidine kinase from Herpes simplex virus type 1 in complex with N-methyl-DHBT | | Descriptor: | N-Methyl-6-(1,3-dihydroxy-isobutyl)thymine, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Perozzo, R, Westermaier, Y, Martic, M, Ametamey, S, Scapozza, L. | | Deposit date: | 2008-10-26 | | Release date: | 2009-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, crystal structure, and in vitro biological evaluation of C-6 pyrimidine derivatives: new lead structures for monitoring gene expression in vivo.
Nucleosides Nucleotides Nucleic Acids, 30, 2011
|
|
1VZT
 
 | | ROLES OF INDIVIDUAL RESIDUES OF ALPHA-1,3 GALACTOSYLTRANSFERASES IN SUBSTRATE BINDING AND CATALYSIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, N-ACETYLLACTOSAMINIDE ALPHA-1,3-GALACTOSYLTRANSFERASE, ... | | Authors: | Zhang, Y, Swaminathan, G.J, Deshpande, A, Natesh, R, Xie, X, Acharya, K.R, Brew, K. | | Deposit date: | 2004-05-26 | | Release date: | 2005-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of Individual Enzyme-Substrate Interactions by Alpha-1,3-Galactosyltransferase in Catalysis and Specificity.
Biochemistry, 42, 2003
|
|
1SBG
 
 | |
1A9M
 
 | | G48H MUTANT OF HIV-1 PROTEASE IN COMPLEX WITH A PEPTIDIC INHIBITOR U-89360E | | Descriptor: | HIV-1 PROTEASE, N-[[1-[N-ACETAMIDYL]-[1-CYCLOHEXYLMETHYL-2-HYDROXY-4-ISOPROPYL]-BUT-4-YL]-CARBONYL]-GLUTAMINYL-ARGINYL-AMIDE | | Authors: | Hong, L, Zhang, X.-J, Foundling, S, Hartsuck, J.A, Tang, J. | | Deposit date: | 1998-04-08 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a G48H mutant of HIV-1 protease explains how glycine-48 replacements produce mutants resistant to inhibitor drugs.
FEBS Lett., 420, 1997
|
|
3MXE
 
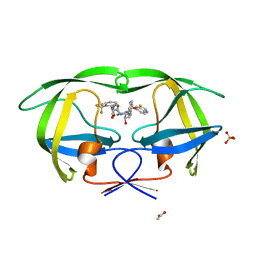 | | Crystal structure of HIV-1 protease inhibitor, KC32 complexed with wild-type protease | | Descriptor: | (5S)-N-{(1S,2R)-3-[(1,3-benzothiazol-6-ylsulfonyl)(2-methylpropyl)amino]-1-benzyl-2-hydroxypropyl}-2-oxo-3-[2-(trifluoromethyl)phenyl]-1,3-oxazolidine-5-carboxamide, ACETATE ION, HIV-1 protease, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2010-05-07 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design, Synthesis, and Structure-Activity Relationship Studies of HIV-1 Protease Inhibitors Incorporating Phenyloxazolidinones.
J.Med.Chem., 53, 2010
|
|
1AJX
 
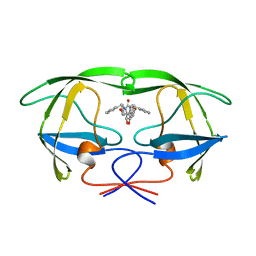 | |
3NJ3
 
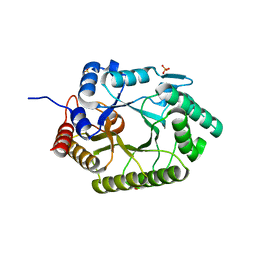 | | Crystal structure of xylanase 10B from Thermotoga petrophila RKU-1 in complex with xylobiose | | Descriptor: | ACETATE ION, Endo-1,4-beta-xylanase, SULFATE ION, ... | | Authors: | Santos, C.R, Meza, A.N, Trindade, D.M, Ruller, R, Squina, F.M, Prade, R.A, Murakami, M.T. | | Deposit date: | 2010-06-16 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Thermal-induced conformational changes in the product release area drive the enzymatic activity of xylanases 10B: Crystal structure, conformational stability and functional characterization of the xylanase 10B from Thermotoga petrophila RKU-1.
Biochem.Biophys.Res.Commun., 403, 2010
|
|
1AID
 
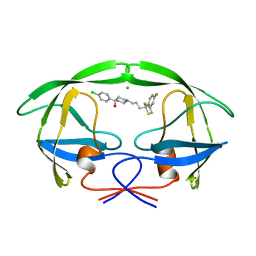 | | STRUCTURE OF A NON-PEPTIDE INHIBITOR COMPLEXED WITH HIV-1 PROTEASE: DEVELOPING A CYCLE OF STRUCTURE-BASED DRUG DESIGN | | Descriptor: | 4-(4-CHLORO-PHENYL)-1-{3-[2-(4-FLUORO-PHENYL)-[1,3]DITHIOLAN-2-YL]-PROPYL}-PIPERIDIN-4-OL, CHLORIDE ION, HUMAN IMMUNODEFICIENCY VIRUS PROTEASE | | Authors: | Rutenber, E.E, Fauman, E.B, Keenan, R.J, Stroud, R.M. | | Deposit date: | 1997-04-16 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a non-peptide inhibitor complexed with HIV-1 protease. Developing a cycle of structure-based drug design.
J.Biol.Chem., 268, 1993
|
|
3NU3
 
 | | Wild Type HIV-1 Protease with Antiviral Drug Amprenavir | | Descriptor: | CHLORIDE ION, GLYCEROL, Protease, ... | | Authors: | Wang, Y.-F, Kovalevsky, A.Y, Weber, I.T. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Amprenavir complexes with HIV-1 protease and its drug-resistant mutants altering hydrophobic clusters.
Febs J., 277, 2010
|
|
1C6Z
 
 | | ALTERNATE BINDING SITE FOR THE P1-P3 GROUP OF A CLASS OF POTENT HIV-1 PROTEASE INHIBITORS AS A RESULT OF CONCERTED STRUCTURAL CHANGE IN 80'S LOOP. | | Descriptor: | (2S)-N-[(2S,3R)-4-[(2S,3S,4aS,8aS)-3-(tert-butylcarbamoyl)-3,4,4a,5,6,7,8,8a-octahydro-1H-isoquinolin-2-yl]-3-hydroxy-1 -phenyl-butan-2-yl]-2-(quinolin-2-ylcarbonylamino)butanediamide, PROTEIN (PROTEASE) | | Authors: | Munshi, S. | | Deposit date: | 1999-12-28 | | Release date: | 2000-12-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An alternate binding site for the P1-P3 group of a class of potent HIV-1 protease inhibitors as a result of concerted structural change in the 80s loop of the protease.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1C70
 
 | |
3ZC5
 
 | | X-ray Structure of c-Met kinase in complex with inhibitor (S)-6-(1-(6- (1-methyl-1H-pyrazol-4-yl)-(1,2,4)triazolo(4,3-b)pyridazin-3-yl)ethyl) quinoline. | | Descriptor: | 6-{(1S)-1-[6-(1-methyl-1H-pyrazol-4-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]ethyl}quinoline, HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | McTigue, M, Grodsky, N, Ryan, K. | | Deposit date: | 2012-11-15 | | Release date: | 2013-11-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Lessons from (S)-6-(1-(6-(1-Methyl-1H-Pyrazol-4-Yl)-[1,2, 4]Triazolo[4,3-B]Pyridazin-3-Yl)Ethyl)Quinoline (Pf-04254644), an Inhibitor of Receptor Tyrosine Kinase C-met with High Protein Kinase Selectivity But Broad Phosphodiesterase Family Inhibition Leading to Myocardial Degeneration in Rats.
J.Med.Chem., 56, 2013
|
|
1BVG
 
 | | HIV-1 PROTEASE-DMP323 COMPLEX IN SOLUTION, NMR MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 PROTEASE, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Yamazaki, T, Hinck, A.P, Wang, Y.-X, Nicholson, L.K, Torchia, D.A, Wingfield, P, Stahl, S.J, Kaufman, J.D, Chang, C, Domaille, P.J, Lam, P.Y.S. | | Deposit date: | 1996-01-16 | | Release date: | 1996-08-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of the HIV-1 protease complexed with DMP323, a novel cyclic urea-type inhibitor, determined by nuclear magnetic resonance spectroscopy.
Protein Sci., 5, 1996
|
|
3VF5
 
 | | Crystal Structure of HIV-1 Protease Mutant I47V with novel P1'-Ligands GRL-02031 | | Descriptor: | (3aS,5R,6aR)-hexahydro-2H-cyclopenta[b]furan-5-yl [(1S,2R)-1-benzyl-2-hydroxy-3-([(4-methoxyphenyl)sulfonyl]{[(2R)-5-oxopyrrolidin-2-yl]methyl}amino)propyl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Yu, X.X, Wang, Y.F, Chang, Y.C.E, Weber, I.T. | | Deposit date: | 2012-01-09 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring.
J.Med.Chem., 55, 2012
|
|
3GI5
 
 | | Crystal structure of protease inhibitor, KB62 in complex with wild type HIV-1 protease | | Descriptor: | (5S)-3-(3-Acetylphenyl)-N-[(1S,2R)-3-[(1,3-benzodioxol-5-ylsulfonyl)(2-methylpropyl)amino]-2-hydroxy-1-(phenylmethyl)pr opyl]-2-oxo-5-oxazolidinecarboxamide, PHOSPHATE ION, Protease | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2009-03-05 | | Release date: | 2010-03-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance.
J.Virol., 84, 2010
|
|
3V81
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) with DNA and the nonnucleoside inhibitor nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, DNA (5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*(MRG)P*CP*GP*CP*CP*(ATM))-3'), DNA (5'-D(*AP*TP*GP*GP*AP*AP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*TP*GP*TP*G)-3'), ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2011-12-22 | | Release date: | 2012-01-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8503 Å) | | Cite: | HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3QIN
 
 | |
3WSJ
 
 | | HTLV-1 protease in complex with the HIV-1 protease inhibitor Indinavir | | Descriptor: | N-[2(R)-HYDROXY-1(S)-INDANYL]-5-[(2(S)-TERTIARY BUTYLAMINOCARBONYL)-4(3-PYRIDYLMETHYL)PIPERAZINO]-4(S)-HYDROXY-2(R)-PHENYLMETHYLPENTANAMIDE, Protease, SULFATE ION, ... | | Authors: | Kuhnert, M, Steuber, H, Diederich, W.E. | | Deposit date: | 2014-03-14 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Structural basis for HTLV-1 protease inhibition by the HIV-1 protease inhibitor indinavir.
J.Med.Chem., 57, 2014
|
|
1T1R
 
 | | Crystal Structure of the Reductoisomerase Complexed with a Bisphosphonate | | Descriptor: | 1-deoxy-D-xylulose 5-phosphate reductoisomerase, SULFATE ION, [(ISOQUINOLIN-1-YLAMINO)-PHOSPHONO-METHYL]-PHOSPHONIC ACID | | Authors: | Yajima, S, Hara, K, Sanders, J.M, Yin, F, Ohsawa, K, Wiesner, J, Jomaa, H, Oldfield, E. | | Deposit date: | 2004-04-17 | | Release date: | 2004-09-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic Structures of Two Bisphosphonate:1-Deoxyxylulose-5-Phosphate Reductoisomerase Complexes
J.Am.Chem.Soc., 126, 2004
|
|
3ST5
 
 | | Crystal structure of wild-type HIV-1 protease with C3-Substituted Hexahydrocyclopentafuranyl Urethane as P2-Ligand, GRL-0489A | | Descriptor: | (3R,3aR,5R,6aR)-3-hydroxyhexahydro-2H-cyclopenta[b]furan-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design of HIV-1 Protease Inhibitors with C3-Substituted Hexahydrocyclopentafuranyl Urethanes as P2-Ligands: Synthesis, Biological Evaluation, and Protein-Ligand X-ray Crystal Structure.
J.Med.Chem., 54, 2011
|
|
3PGT
 
 | | CRYSTAL STRUCTURE OF HGSTP1-1[I104] COMPLEXED WITH THE GSH CONJUGATE OF (+)-ANTI-BPDE | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-AMINO-4-[1-(CARBOXYMETHYL-CARBAMOYL)-2-(9-HYDROXY-7,8-DIOXO-7,8,9,10-TETRAHYDRO-BENZO[DEF]CHRYSEN-10-YLSULFANYL)-ETHYLCARBAMOYL]-BUTYRIC ACID, PROTEIN (GLUTATHIONE S-TRANSFERASE), ... | | Authors: | Ji, X, Xiao, B. | | Deposit date: | 1999-03-22 | | Release date: | 1999-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure and function of residue 104 and water molecules in the xenobiotic substrate-binding site in human glutathione S-transferase P1-1.
Biochemistry, 38, 1999
|
|
3PZM
 
 | | Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Mannan endo-1,4-beta-mannosidase. Glycosyl Hydrolase family 5 | | Authors: | Santos, C.R, Meza, A.N, Paiva, J.H, Silva, J.C, Ruller, R, Prade, R.A, Squina, F.M, Murakami, M.T. | | Deposit date: | 2010-12-14 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural characterization of a novel hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1
To be Published
|
|
1RTH
 
 | | HIGH RESOLUTION STRUCTURES OF HIV-1 RT FROM FOUR RT-INHIBITOR COMPLEXES | | Descriptor: | 6,11-DIHYDRO-11-ETHYL-6-METHYL-9-NITRO-5H-PYRIDO[2,3-B][1,5]BENZODIAZEPIN-5-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Esnouf, R, Garman, E, Somers, D, Ross, C, Kirby, I, Keeling, J, Darby, G, Jones, Y, Stuart, D, Stammers, D. | | Deposit date: | 1995-05-03 | | Release date: | 1996-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High resolution structures of HIV-1 RT from four RT-inhibitor complexes.
Nat.Struct.Biol., 2, 1995
|
|
1REV
 
 | | HIV-1 REVERSE TRANSCRIPTASE | | Descriptor: | 4-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, HIV-1 REVERSE TRANSCRIPTASE, MAGNESIUM ION | | Authors: | Ren, J, Esnouf, R, Hopkins, A, Ross, C, Jones, Y, Stammers, D, Stuart, D. | | Deposit date: | 1995-09-17 | | Release date: | 1996-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of HIV-1 reverse transcriptase complexed with 9-chloro-TIBO: lessons for inhibitor design.
Structure, 3, 1995
|
|
2E40
 
 | | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium in complex with gluconolactone | | Descriptor: | Beta-glucosidase, D-glucono-1,5-lactone | | Authors: | Nijikken, Y, Tsukada, T, Igarashi, K, Samejima, M, Fushinobu, S. | | Deposit date: | 2006-12-01 | | Release date: | 2007-03-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of intracellular family 1 beta-glucosidase BGL1A from the basidiomycete Phanerochaete chrysosporium
Febs Lett., 581, 2007
|
|
